
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,109,268 – 12,109,406 |
| Length | 138 |
| Max. P | 0.942138 |

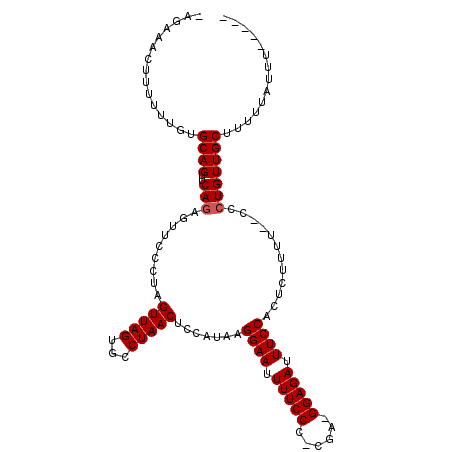
| Location | 12,109,268 – 12,109,373 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12109268 105 + 22224390 -AGAAACUUUUUUACGCAGUUCAGAGCUCCCUAGUUAGUGCCUAACUCCAUAAGGAAUUUUCCCCCGA-GGAGAUUUCCACUCUUUUUCCCCUGUUGCUUUUUAUUU----- -..............((((..(((........((((((...))))))......((((.(((((.....-))))).))))............))))))).........----- ( -18.30) >DroSec_CAF1 21479 104 + 1 -AGAAACUUUUUUGUGCAGUUCAGAGUUCCCUAGUUAGUGCCUAACUCCAUAAGGAAUUUUCCCCCCAGGGAGAUUUCCACUCUUUU--CCCUGUUGCUUUUUAUUU----- -..............((((...(((((.....((((((...))))))......((((.((((((....)))))).)))))))))...--..))))............----- ( -22.90) >DroEre_CAF1 32998 102 + 1 -AGAAACUUUUUUGUGCAGCUCACAGUUUCCUAGUUAGUGCCUAACUCCAUAGGGAAUUUUCCC-CGA-GGAGAUUUCCACUCUUUU--CCCUGUUGCUUUUUAUUU----- -..............(((((....(((((((((...(((.....)))...))))))))).....-.((-(((((.......))))))--)...))))).........----- ( -21.30) >DroYak_CAF1 34220 110 + 1 CAAAAACUUUUUUGUGCAGUUCAGAGUUCCCUUGUUAGUGCCUAACUCCAUAAGGAAUUUUCCC-CGA-GGAGAUUUCCACUCUUUUUCCCCUGUUGCUUUUUUUUUUUAUU .(((((.........((((...(((((......(((((...))))).......((((.(((((.-...-))))).))))))))).......)))).........)))))... ( -18.17) >consensus _AGAAACUUUUUUGUGCAGUUCAGAGUUCCCUAGUUAGUGCCUAACUCCAUAAGGAAUUUUCCC_CGA_GGAGAUUUCCACUCUUUU__CCCUGUUGCUUUUUAUUU_____ ...............((((..(((.........(((((...))))).......((((.(((((......))))).))))............))))))).............. (-16.83 = -17.07 + 0.25)


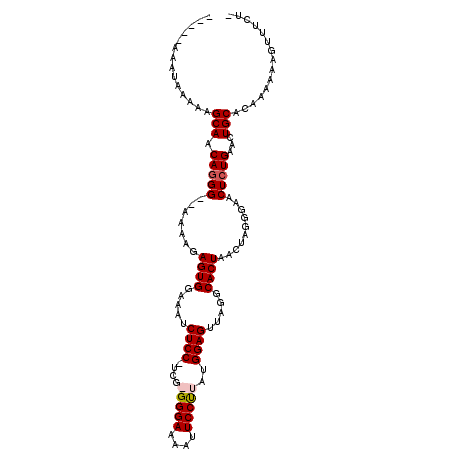
| Location | 12,109,268 – 12,109,373 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12109268 105 - 22224390 -----AAAUAAAAAGCAACAGGGGAAAAAGAGUGGAAAUCUCC-UCGGGGGAAAAUUCCUUAUGGAGUUAGGCACUAACUAGGGAGCUCUGAACUGCGUAAAAAAGUUUCU- -----.........(((.(((((.......((((.....((((-...(((((....)))))..)))).....))))..........)))))...)))..............- ( -21.83) >DroSec_CAF1 21479 104 - 1 -----AAAUAAAAAGCAACAGGG--AAAAGAGUGGAAAUCUCCCUGGGGGGAAAAUUCCUUAUGGAGUUAGGCACUAACUAGGGAACUCUGAACUGCACAAAAAAGUUUCU- -----.........(((.(((((--....((.......))(((((((..((..((((((....)))))).....))..))))))).)))))...)))..............- ( -26.00) >DroEre_CAF1 32998 102 - 1 -----AAAUAAAAAGCAACAGGG--AAAAGAGUGGAAAUCUCC-UCG-GGGAAAAUUCCCUAUGGAGUUAGGCACUAACUAGGAAACUGUGAGCUGCACAAAAAAGUUUCU- -----........(((.((((..--.....((((.....((((-..(-((((....)))))..)))).....))))..........))))..)))................- ( -24.53) >DroYak_CAF1 34220 110 - 1 AAUAAAAAAAAAAAGCAACAGGGGAAAAAGAGUGGAAAUCUCC-UCG-GGGAAAAUUCCUUAUGGAGUUAGGCACUAACAAGGGAACUCUGAACUGCACAAAAAAGUUUUUG ..............(((.(((((.......((((.....((((-..(-((((....)))))..)))).....))))..........)))))...)))............... ( -22.93) >consensus _____AAAUAAAAAGCAACAGGG__AAAAGAGUGGAAAUCUCC_UCG_GGGAAAAUUCCUUAUGGAGUUAGGCACUAACUAGGGAACUCUGAACUGCACAAAAAAGUUUCU_ ..............(((.(((((.......((((.....((((....(((((....)))))..)))).....))))..........)))))...)))............... (-18.64 = -19.20 + 0.56)

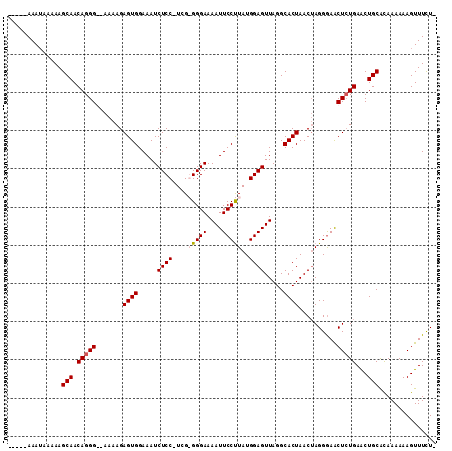
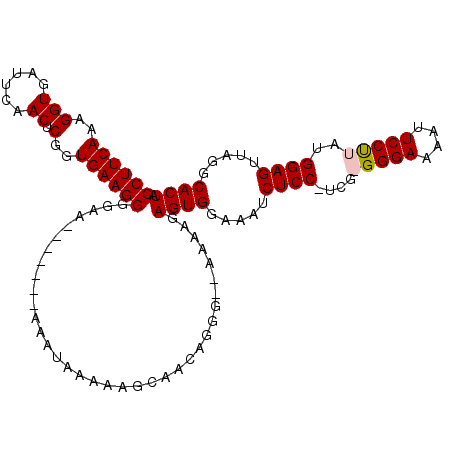
| Location | 12,109,302 – 12,109,406 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -22.49 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12109302 104 - 22224390 CCUUCAAAGGUGAUUCAACUCGGUGAAGGGGAA-------AAAUAAAAAGCAACAGGGGAAAAAGAGUGGAAAUCUCC-UCGGGGGAAAAUUCCUUAUGGAGUUAGGCACUA ((((((..(((......)).)..))))))....-------.........................((((.....((((-...(((((....)))))..)))).....)))). ( -24.80) >DroSec_CAF1 21513 103 - 1 CCUUCAAAGGUGAUUCAACUCGGUGAAGGGGAA-------AAAUAAAAAGCAACAGGG--AAAAGAGUGGAAAUCUCCCUGGGGGGAAAAUUCCUUAUGGAGUUAGGCACUA ((((((..(((......)).)..))))))....-------.........((..(((((--(...((.......)))))))).......((((((....))))))..)).... ( -26.80) >DroEre_CAF1 33032 101 - 1 CCUUCAAAGGUGAUUCAACUCGGUGAAGGGGAA-------AAAUAAAAAGCAACAGGG--AAAAGAGUGGAAAUCUCC-UCG-GGGAAAAUUCCCUAUGGAGUUAGGCACUA ((((((..(((......)).)..))))))....-------..................--.....((((.....((((-..(-((((....)))))..)))).....)))). ( -27.60) >DroYak_CAF1 34255 110 - 1 CCUUCAAAGGUGAUUCAACUCGGUGAAGGGGAAUAAAUAAAAAAAAAAAGCAACAGGGGAAAAAGAGUGGAAAUCUCC-UCG-GGGAAAAUUCCUUAUGGAGUUAGGCACUA ((((((..(((......)).)..))))))....................................((((.....((((-..(-((((....)))))..)))).....)))). ( -24.80) >consensus CCUUCAAAGGUGAUUCAACUCGGUGAAGGGGAA_______AAAUAAAAAGCAACAGGG__AAAAGAGUGGAAAUCUCC_UCG_GGGAAAAUUCCUUAUGGAGUUAGGCACUA ((((((..(((......)).)..))))))....................................((((.....((((....(((((....)))))..)))).....)))). (-22.49 = -22.80 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:51 2006