
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,098,184 – 12,098,318 |
| Length | 134 |
| Max. P | 0.987662 |

| Location | 12,098,184 – 12,098,287 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12098184 103 - 22224390 CGAGCAUCAAAAAAGAAGGAAAA-------------UGUAUGUGUGUGUGUGCGCGGCACACAUAUCAGCAUAAGUCA----GGAUGCGGAUAUACAUACAUGUGUACAUGUGAGUAUCA ...(((((..........((...-------------..(((((((((((((((...))))))))))..)))))..)).----.))))).(((((...((((((....)))))).))))). ( -30.90) >DroSec_CAF1 10550 99 - 1 GGAGCAUCAAAAAGGAAGAAAAA-------------UGUAUGUGUGUGU----GCGGCACACAUAUCAGCAUAAGUCAGGCAGGAUGCGGAUAUA----CAUGUGUACAUGUGAGUAUCA ...(((((......((.......-------------((.(((((((((.----....))))))))))).......))......))))).((((((----((((....)))))..))))). ( -30.14) >DroEre_CAF1 22165 91 - 1 GGAGCAUCAAAAAAGAAGGAAAA-------------UGUGU--GUGUGUGUGGGCGGCACACAUAUCAGCAUAAGUCA----GGA------UGUA----CAUGUGUACAUGUGAGUAUCA .((...(((.........((..(-------------(((((--((((((((.....))))))))))..))))...)).----..(------((((----(....)))))).)))...)). ( -24.10) >DroYak_CAF1 22639 110 - 1 GGAGCAUCAAAAAAGAGGGAAUACAUACAUUUUCCUUGUGG--GUGUGUGUGUGUGACACACAUAUCAGCAUAAGUCA----GGAUGUGGAUGUA----CAUGUGUACAUGUGAGUAUCA ...(((((......(((((((.........)))))))((.(--(((((((((.....)))))))))).))........----.))))).((((((----((((....))))))..)))). ( -33.90) >consensus GGAGCAUCAAAAAAGAAGGAAAA_____________UGUAU__GUGUGUGUG_GCGGCACACAUAUCAGCAUAAGUCA____GGAUGCGGAUAUA____CAUGUGUACAUGUGAGUAUCA .((...(((...........................(((....((((((((.....))))))))....)))............................((((....)))))))...)). (-15.38 = -15.75 + 0.38)

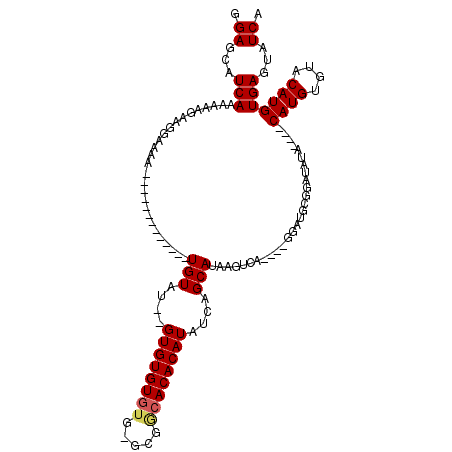

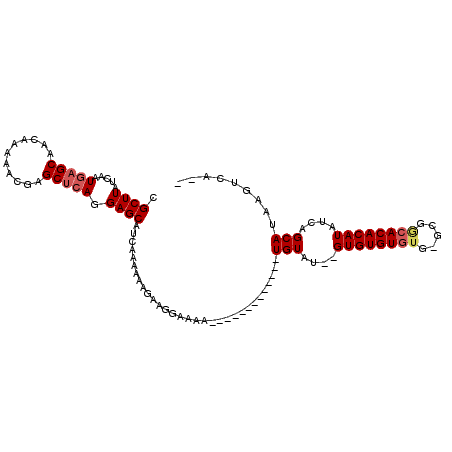
| Location | 12,098,222 – 12,098,318 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12098222 96 - 22224390 CGCUUAUCAAUGAGCAACAAAAACGAGCUCACGAGCAUCAAAAAAGAAGGAAAA-------------UGUAUGUGUGUGUGUGCGCGGCACACAUAUCAGCAUAAGUCA-- .((((.....(((((...........))))).)))).............((...-------------..(((((((((((((((...))))))))))..)))))..)).-- ( -24.90) >DroSec_CAF1 10586 94 - 1 CGCUUAUCAAUGAGCAACAAAAACGAGCUCAGGAGCAUCAAAAAGGAAGAAAAA-------------UGUAUGUGUGUGU----GCGGCACACAUAUCAGCAUAAGUCAGG .((((.....(((((...........))))).))))............((...(-------------((((((((((((.----....)))))))))..))))...))... ( -25.20) >DroEre_CAF1 22193 94 - 1 CGCUUAUCAAUGAGCAACAAAAACGAGCCGAGGAGCAUCAAAAAAGAAGGAAAA-------------UGUGU--GUGUGUGUGGGCGGCACACAUAUCAGCAUAAGUCA-- .(((((....))))).........((((......)).))..........((..(-------------(((((--((((((((.....))))))))))..))))...)).-- ( -21.30) >DroYak_CAF1 22673 107 - 1 CGCUUAUCAAUGAGCAACAGAAACGAGCUCAGGAGCAUCAAAAAAGAGGGAAUACAUACAUUUUCCUUGUGG--GUGUGUGUGUGUGACACACAUAUCAGCAUAAGUCA-- .((((.....(((((...........))))).)))).........(((((((.........)))))))((.(--(((((((((.....)))))))))).))........-- ( -31.50) >consensus CGCUUAUCAAUGAGCAACAAAAACGAGCUCAGGAGCAUCAAAAAAGAAGGAAAA_____________UGUAU__GUGUGUGUG_GCGGCACACAUAUCAGCAUAAGUCA__ .((((.....(((((...........))))).))))...............................(((....((((((((.....))))))))....)))......... (-17.48 = -18.35 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:42 2006