| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,092,271 – 12,092,371 |
| Length | 100 |
| Max. P | 0.999914 |

| Location | 12,092,271 – 12,092,371 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.67 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
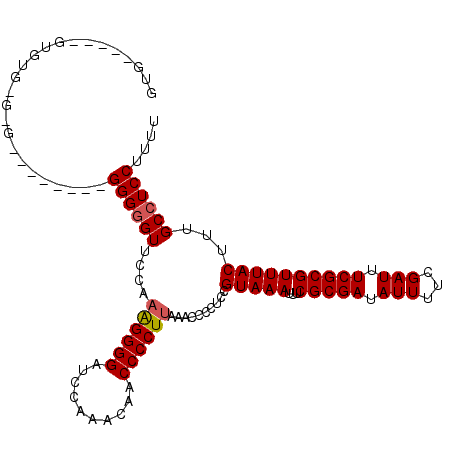
>X_DroMel_CAF1 12092271 100 + 22224390 GUAU----GUGUGGGAG--------GGGUGU-UUCAGGGGGUU-CCAACAACCCCUUAAACCCCUCCGUAAAUUUCGCGAUAUUUUCGAUUUCGCGUUUACUUUGCCUCCUUUU ....----.....((((--------((((..-...((((((((-.....))))))))..))))))))(((((...(((((.(((...))).))))))))))............. ( -36.20) >DroEre_CAF1 16350 92 + 1 GAG----------------------GGGGGUUCCAAAGGGGAUCCAAACCACCCCUUAAACCCCUCCGUAAAUUUCGCGAUAUUUUCGAUUUCGCGUUUACUUUGCCUCCUUUU (((----------------------(((((((...((((((..........)))))).))))))...(((((...(((((.(((...))).))))))))))....))))..... ( -31.50) >DroYak_CAF1 16712 114 + 1 GUGAGUGAGUGUGAGCGGGGUUCCAGGGGGUUCCAAAGGGGGUCUUAACAACCCCUGAAACCCAUCCGUAAAUUUCGCGAUAUUUUCGAUUUCGCGUUUACUUUGCCUCCUUUU ..(((((((((((((((..(((...(((((((....(((((..........)))))..)))))..))...)))..)))((.....))....))))))))))))........... ( -37.20) >consensus GUG_____GUGUG_G_G________GGGGGUUCCAAAGGGGAUCCAAACAACCCCUUAAACCCCUCCGUAAAUUUCGCGAUAUUUUCGAUUUCGCGUUUACUUUGCCUCCUUUU .........................((((((....((((((..........))))))..........(((((...(((((.(((...))).))))))))))...)))))).... (-23.25 = -23.70 + 0.45)


| Location | 12,092,271 – 12,092,371 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -20.22 |
| Energy contribution | -21.33 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
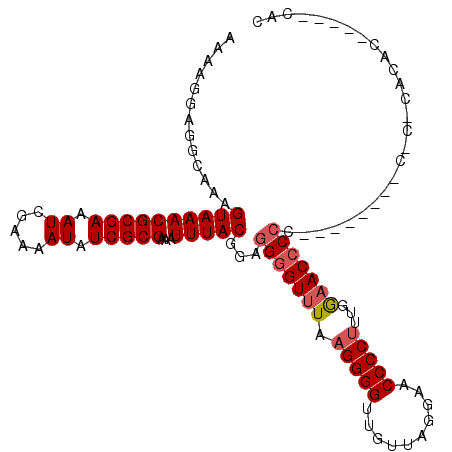
>X_DroMel_CAF1 12092271 100 - 22224390 AAAAGGAGGCAAAGUAAACGCGAAAUCGAAAAUAUCGCGAAAUUUACGGAGGGGUUUAAGGGGUUGUUGG-AACCCCCUGAA-ACACCC--------CUCCCACAC----AUAC .............((((((((((.((.....)).)))))...)))))(((((((((((.((((((.....-)))))).))).-..))))--------)))).....----.... ( -34.20) >DroEre_CAF1 16350 92 - 1 AAAAGGAGGCAAAGUAAACGCGAAAUCGAAAAUAUCGCGAAAUUUACGGAGGGGUUUAAGGGGUGGUUUGGAUCCCCUUUGGAACCCCC----------------------CUC .....((((....((((((((((.((.....)).)))))...)))))...(((((((((((((.(((....))))))))).))))))))----------------------))) ( -32.00) >DroYak_CAF1 16712 114 - 1 AAAAGGAGGCAAAGUAAACGCGAAAUCGAAAAUAUCGCGAAAUUUACGGAUGGGUUUCAGGGGUUGUUAAGACCCCCUUUGGAACCCCCUGGAACCCCGCUCACACUCACUCAC ....(..(((...((((((((((.((.....)).)))))...)))))....((((((((((((((.(.(((.....))).).)).)))))))))))).)))..).......... ( -37.50) >consensus AAAAGGAGGCAAAGUAAACGCGAAAUCGAAAAUAUCGCGAAAUUUACGGAGGGGUUUAAGGGGUUGUUAGGAACCCCUUUGGAACCCCC________C_C_CACAC_____CAC .............((((((((((.((.....)).)))))...)))))...(((((((.(((((..........)))))...))))))).......................... (-20.22 = -21.33 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:37 2006