| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,358,594 – 1,358,694 |
| Length | 100 |
| Max. P | 0.565690 |

| Location | 1,358,594 – 1,358,694 |
|---|---|
| Length | 100 |
| Sequences | 6 |
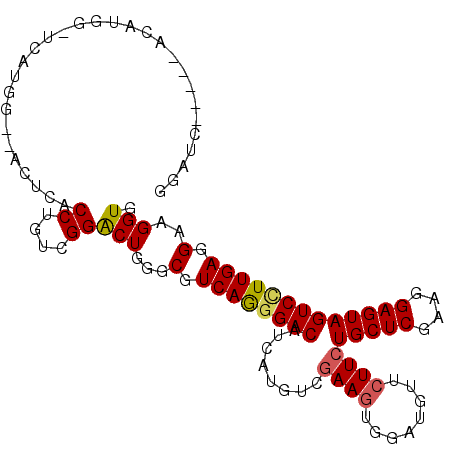
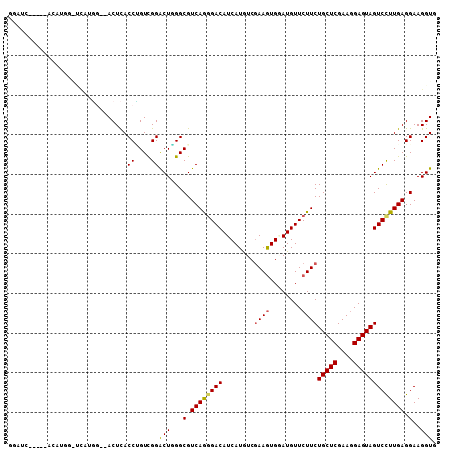
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1358594 100 + 22224390 GGUGCU----UUG----UGUAGGAAACUCACCCGUCGGGCUGGGCGUCAGCGACAUCAUGUCGAAGUGGAUGUUCUUCUGCUCGAAGGAGUAGUCCUUGAGGAAGGUC ...(((----((.----(..((((.((((.....((((((.(((((((..((((.....)))).....)))))))....))))))..))))..))))...).))))). ( -34.90) >DroPse_CAF1 28899 102 + 1 GGAUCC----CCAUGGAUCAUGG--ACUCACCUGUCGGACUGGGCGUCAGGGACAUCAUGUCGAAGUGGAUGUUCUUCUGCUCGAAGGAGUAGUCUUUGAGGAAGGUU .(((((----....)))))...(--((......))).((((...(.((((((((.((...((((.(..((......))..)))))..))...)))))))).)..)))) ( -34.30) >DroGri_CAF1 29244 104 + 1 GAAUG-UAAUAAUUGGGAU-UU--CACUUACCGGUCGGACUCGGCGUCAGGGACAUCAUAUCGAAGUGAAUGUUCUUCUGCUCGAAUGAGUAGUCCUUGAGGAAGGUG .....-.............-..--(((((.((.((((....)))).((((((((.((((.((((.(((((.....))).))))))))))...)))))))))).))))) ( -29.90) >DroMoj_CAF1 34147 96 + 1 ----------ACUUGGGCUAUU--CACUUACCAGUUGGACUUGGCGUCAAGGACAUCAUGUCGAAAUGUAUGUUCUUCUGCUCGAAGGAGUAGUCCUUGAGGAAGGUG ----------.(..((((((((--(.(((.((....))....((((..(((((((((((......))).)))))))).))))..))))))))))))..)......... ( -29.00) >DroAna_CAF1 19285 98 + 1 UAUACU----AUA----GCAAGG--ACUCACCUGUUGGACUCGGCGUCAGGGACAUCAUGUCAAAGUGUAUGUUCUUCUGCUCGAAGGAGUAGUCCUUGAGGAAGGUG ..((((----.(.----.(((((--((((.(((.......((((((..(((((((((((......))).)))))))).))).)))))).).))))))))...).)))) ( -29.81) >DroPer_CAF1 29727 102 + 1 GGAUCC----CCAUGGAUCAUGG--ACUCACCUGUCGGACUGGGCGUCAGGGACAUCAUGUCGAAGUGGAUGUUCUUCUGCUCGAAGGAGUAGUCUUUGAGGAAGGUU .(((((----....)))))...(--((......))).((((...(.((((((((.((...((((.(..((......))..)))))..))...)))))))).)..)))) ( -34.30) >consensus GGAUC_____ACAUGG_UCAUGG__ACUCACCUGUCGGACUGGGCGUCAGGGACAUCAUGUCGAAGUGGAUGUUCUUCUGCUCGAAGGAGUAGUCCUUGAGGAAGGUG ..............................((....))(((...(.((((((((........((((........))))(((((....))))))))))))).)..))). (-23.64 = -23.70 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:06 2006