
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,085,166 – 12,085,279 |
| Length | 113 |
| Max. P | 0.999909 |

| Location | 12,085,166 – 12,085,279 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -23.45 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12085166 113 + 22224390 ----GAAAUUGUGAGGGGGGUGGAGCUGCGGGAGGGAAAACCACCCA-GGAAAACCACCCAGCACUCCGAGGCUGCACAACAAAAACCACAUUCAUAGAUAAUGUGCAGCUUGGGUUU ----......(((....((((((..((..(((.((.....)).))).-))....))))))..))).((.((((((((((.......................)))))))))).))... ( -42.00) >DroSec_CAF1 20922 110 + 1 GGGGGGAAAUGUAAAGGGAGAGG-------GCUGGGAAAACCACCCA-GGAAAAGCACCCAGCCCUCCGAGGCUGCACAACAAAAACCACAUUUAUAGAUAAUGUGCAGCUUGGGUUU ...................((((-------((((((....((.....-)).......))))))))))(.((((((((((.......................)))))))))).).... ( -37.00) >DroAna_CAF1 19236 102 + 1 ----GAA--------GUGGGUGGAG-UGCACCACCGCCCACCACCCACCGAAAACCACCCA---CUCGGAGGCUGCACAACAAAAACCACAUUCAUAGAUAAUGUGCAGCCUGGGUUU ----...--------((((((((.(-.....).)))))))).((((.((((..........---.))))((((((((((.......................)))))))))))))).. ( -39.80) >consensus ____GAAA_UGU_A_GGGGGUGGAG_UGC_GCAGGGAAAACCACCCA_GGAAAACCACCCAGC_CUCCGAGGCUGCACAACAAAAACCACAUUCAUAGAUAAUGUGCAGCUUGGGUUU .................((((((.................))))))..........(((((.........(((((((((.......................)))))))))))))).. (-23.45 = -23.90 + 0.45)

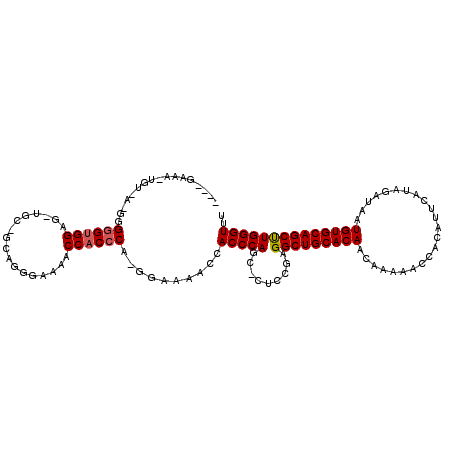

| Location | 12,085,166 – 12,085,279 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -25.88 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12085166 113 - 22224390 AAACCCAAGCUGCACAUUAUCUAUGAAUGUGGUUUUUGUUGUGCAGCCUCGGAGUGCUGGGUGGUUUUCC-UGGGUGGUUUUCCCUCCCGCAGCUCCACCCCCCUCACAAUUUC---- ...((...((((((((.((.((((....))))....)).))))))))...)).(((..((((((......-((((.((.....)).)))).....))))))....)))......---- ( -37.90) >DroSec_CAF1 20922 110 - 1 AAACCCAAGCUGCACAUUAUCUAUAAAUGUGGUUUUUGUUGUGCAGCCUCGGAGGGCUGGGUGCUUUUCC-UGGGUGGUUUUCCCAGC-------CCUCUCCCUUUACAUUUCCCCCC ........((((((((.((.((((....))))....)).))))))))...(((((((((((.(((...(.-...).)))...))))))-------))))).................. ( -39.70) >DroAna_CAF1 19236 102 - 1 AAACCCAGGCUGCACAUUAUCUAUGAAUGUGGUUUUUGUUGUGCAGCCUCCGAG---UGGGUGGUUUUCGGUGGGUGGUGGGCGGUGGUGCA-CUCCACCCAC--------UUC---- ..((((((((((((((.((.((((....))))....)).))))))))).(((((---.........))))))))))((((((.((.(.....-).)).)))))--------)..---- ( -43.40) >consensus AAACCCAAGCUGCACAUUAUCUAUGAAUGUGGUUUUUGUUGUGCAGCCUCGGAG_GCUGGGUGGUUUUCC_UGGGUGGUUUUCCCUGC_GCA_CUCCACCCCC_U_ACA_UUUC____ ..(((((.((((((((.((.((((....))))....)).))))))))..........)))))..........((((((.................))))))................. (-25.88 = -26.00 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:32 2006