| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,084,390 – 12,084,483 |
| Length | 93 |
| Max. P | 0.999961 |

| Location | 12,084,390 – 12,084,483 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.30 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.62 |
| SVM decision value | 4.92 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12084390 93 + 22224390 GCCAACAGAAAUUUGUUGGGCAAUUUCCAAGCCAGCAAAUCUCUGCAAUGCUGGUUGCAG-GCAACCCAACCAGCUAAGCCACCCAUUUCACCC----------------- ((...((((.(((((((((............))))))))).))))....((((((((..(-....).))))))))...))..............----------------- ( -29.00) >DroSec_CAF1 20105 92 + 1 GCCAACAGAAAUUUGUUGGGCAAUUUCCAAGCCAACAAAUCUCUGCAAUGCUGG-UGCAG-GCAACCCAACCAGCUAAGCCACCCAUUUCACCA----------------- ((...((((.(((((((((............))))))))).))))....(((((-(...(-....)...))))))...))..............----------------- ( -26.60) >DroMoj_CAF1 34503 102 + 1 GCCAACAGAAAUUUGUUGGGCAAUUUCCAAGCCAACAAAUCUCUGCAAAGCUGC-CGCACUGCAACACUGG--GCUAACAC--CCACCCC----GGCCCCACCCGCACUCA (((...(((.(((((((((............))))))))).)))(((..((...-.))..))).....(((--(......)--)))....----))).............. ( -25.00) >DroAna_CAF1 18194 109 + 1 GCCAACAGAAAUUUGUUGGGCAAUUUCCAAGCCAACAAAUCCCUGCAAUGUCGG-UGCAG-GGAAGGCCUCCAACUAUUCCCGACAUCCCCCCAUAGCCCACCCACUAGAU (((........((((((((............))))))))((((((((.......-)))))-))).)))........................................... ( -26.00) >consensus GCCAACAGAAAUUUGUUGGGCAAUUUCCAAGCCAACAAAUCUCUGCAAUGCUGG_UGCAG_GCAACCCAACCAGCUAAGCCACCCAUCCCACCA_________________ .....((((.(((((((((............))))))))).))))....(((((.((..........)).))))).................................... (-16.44 = -17.00 + 0.56)

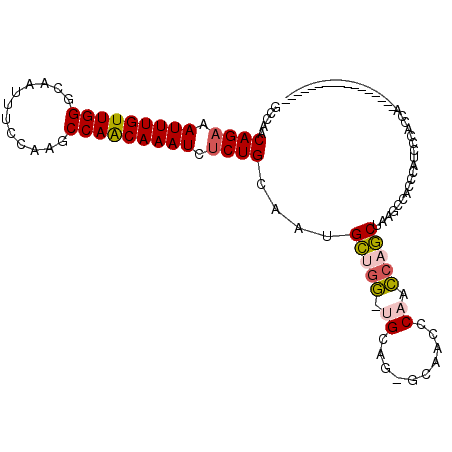
| Location | 12,084,390 – 12,084,483 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.30 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.44 |
| SVM RNA-class probability | 0.999898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12084390 93 - 22224390 -----------------GGGUGAAAUGGGUGGCUUAGCUGGUUGGGUUGC-CUGCAACCAGCAUUGCAGAGAUUUGCUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGC -----------------.(((....((((..((((((....))))))..)-)))..))).((...(((((((((((..(((.........)))...)))))))))))..)) ( -33.90) >DroSec_CAF1 20105 92 - 1 -----------------UGGUGAAAUGGGUGGCUUAGCUGGUUGGGUUGC-CUGCA-CCAGCAUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGC -----------------(((((....(((..((((((....))))))..)-)).))-)))((...((((((((((((((((.........).)))))))))))))))..)) ( -41.10) >DroMoj_CAF1 34503 102 - 1 UGAGUGCGGGUGGGGCC----GGGGUGG--GUGUUAGC--CCAGUGUUGCAGUGCG-GCAGCUUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGC ((.(..(..(..(.((.----.((((..--......))--)).)).)..).)..).-.))(((..((((((((((((((((.........).))))))))))))))).))) ( -38.70) >DroAna_CAF1 18194 109 - 1 AUCUAGUGGGUGGGCUAUGGGGGGAUGUCGGGAAUAGUUGGAGGCCUUCC-CUGCA-CCGACAUUGCAGGGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGC .....((.((((.(....((((((.(.((.((.....)).)).).)))))-)).))-)).))...((((..((((((((((.........).)))))))))..)))).... ( -36.90) >consensus _________________UGGUGAAAUGGGUGGCUUAGCUGGUUGGGUUGC_CUGCA_CCAGCAUUGCAGAGAUUUGUUGGCUUGGAAAUUGCCCAACAAAUUUCUGUUGGC ....................................(((((.(((........))).)))))...((((((((((((((((.........).))))))))))))))).... (-24.17 = -24.80 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:30 2006