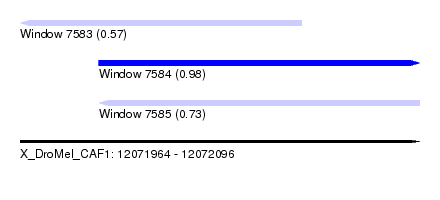
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,071,964 – 12,072,096 |
| Length | 132 |
| Max. P | 0.981223 |

| Location | 12,071,964 – 12,072,057 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.45 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -12.26 |
| Energy contribution | -13.12 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12071964 93 - 22224390 CCAAACAACAAGAUGUCAUAUCAUC--GGAAUGG-CAGUCUCAUUACGAGACUUUGUUC----------AUCGCCUUUUUGUUCG-UUUCCCUUUU--CGUUGUGUCUU----------- ....(((((..((((......))))--.(((..(-.((((((.....)))))))..)))----------................-..........--.))))).....----------- ( -21.00) >DroSec_CAF1 7846 95 - 1 CCAAACAACAAGAUGUCAUAUCAUC--GGAAUGG-CAGUCUCAUUACGAGACUUUGUUC----------AUCGCCUUUUUGUUCG-CUUCCCUUUUUUUGUUGUGUCUU----------- ....(((((((((((......))))--.(((..(-.((((((.....)))))))..)))----------................-...........))))))).....----------- ( -22.50) >DroSim_CAF1 7835 95 - 1 CCAAACAACAAGAUGUCAUAUCAUC--GGAAUGG-CAGUCUCAUUACAAAACUUUGUUC----------AUCUCCUUUUUGUUCC-CUUCCCUUUUUCUGUUGUGUCUU----------- ....((((((.((((......))))--((((..(-.((.......((((....))))..----------.....))..)..))))-............)))))).....----------- ( -15.04) >DroEre_CAF1 7367 93 - 1 CCAAACAACAAGAUGUCAUAUCAUC--GGAAUGG-CAGUCUCAUUACGAGACUUUGUUC----------AUCGCCUUUUUGUUCG-UUUCCCUUUU--UGUUGUGUCUU----------- ....(((((((((((......))))--.(((..(-.((((((.....)))))))..)))----------................-.........)--)))))).....----------- ( -22.50) >DroYak_CAF1 8249 91 - 1 CCAAACAACAAGAUGUCAUAUCAUC--GGAAUGG-CAGUCUCAUUACGAGACUUUGUUC----------GUCGCCUUUUUGUUCG-UUUCCCUUUU--UGUUG--UCUU----------- ....(((((((((((......))))--.(((..(-.((((((.....)))))))..)))----------................-.........)--)))))--)...----------- ( -21.90) >DroAna_CAF1 8253 120 - 1 CCGAACAACAAGAUGUCAUAUCAUCUCCGAAUGGUUACUCUCAUUACGGGACUUUGUUGCCUCCUUUUGUUGGCCAUUUUGUUUCAUUUCUGUUUUGUUGUUGUUUUCUUCGUUACUCUC ..(((((((((((((......)))).((((((((......))))).)))(((...((.(((..(....)..))).))...)))..............))))))))).............. ( -22.80) >consensus CCAAACAACAAGAUGUCAUAUCAUC__GGAAUGG_CAGUCUCAUUACGAGACUUUGUUC__________AUCGCCUUUUUGUUCG_UUUCCCUUUU__UGUUGUGUCUU___________ ....((((((.((((......))))...(((..(..((((((.....)))))))..))).......................................))))))................ (-12.26 = -13.12 + 0.86)



| Location | 12,071,990 – 12,072,096 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.06 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12071990 106 + 22224390 AAAAAGGCGAUGAACAAAGUCUCGUAAUGAGACUGCCAUUCCGAUGAUAUGACAUCUUGUUGUUUGGGUCCACUCGACUGGAGGACUCAAGGACCGAAUGGAGCCA .....(((.........((((((.....)))))).((((((.((((......))))..(((.(((((((((.((.....)).)))))))))))).)))))).))). ( -35.70) >DroSec_CAF1 7874 106 + 1 AAAAAGGCGAUGAACAAAGUCUCGUAAUGAGACUGCCAUUCCGAUGAUAUGACAUCUUGUUGUUUGGGUCCACUCGACUGGAGGACUCAAGGACCAAAAGGAGCCA .....(((.........((((((.....)))))).....(((((((......))))(((.(.(((((((((.((.....)).))))))))).).)))..)))))). ( -33.30) >DroSim_CAF1 7863 106 + 1 AAAAAGGAGAUGAACAAAGUUUUGUAAUGAGACUGCCAUUCCGAUGAUAUGACAUCUUGUUGUUUGGGUCCACUAGAUUGGAGGACUCAAGGACCAAAAGGAGCCA .....((..........((((((.....))))))....((((((((......))))(((.(.(((((((((.((.....)).))))))))).).)))..)))))). ( -26.10) >DroEre_CAF1 7393 106 + 1 AAAAAGGCGAUGAACAAAGUCUCGUAAUGAGACUGCCAUUCCGAUGAUAUGACAUCUUGUUGUUUGGGUCCACUCGACUGGCGGACUCGAGGACCAAAGACAACCA .....((.((((.....((((((.....))))))..))))))((((......))))..(((((((.(((((..((((.........)))))))))..))))))).. ( -30.10) >DroYak_CAF1 8273 106 + 1 AAAAAGGCGACGAACAAAGUCUCGUAAUGAGACUGCCAUUCCGAUGAUAUGACAUCUUGUUGUUUGGGUCCACUCGACUGGAGGGCUCAAGGACCAAAAAGAACCA .....((...(((((((((((((.....))))))........((((......))))...)))))))(((((.(((.....))).......)))))........)). ( -27.20) >consensus AAAAAGGCGAUGAACAAAGUCUCGUAAUGAGACUGCCAUUCCGAUGAUAUGACAUCUUGUUGUUUGGGUCCACUCGACUGGAGGACUCAAGGACCAAAAGGAGCCA .....((.((((.....((((((.....))))))..))))))((((......))))(((.(.(((((((((.((.....)).))))))))).).)))......... (-26.66 = -26.06 + -0.60)



| Location | 12,071,990 – 12,072,096 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -23.54 |
| Energy contribution | -24.06 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12071990 106 - 22224390 UGGCUCCAUUCGGUCCUUGAGUCCUCCAGUCGAGUGGACCCAAACAACAAGAUGUCAUAUCAUCGGAAUGGCAGUCUCAUUACGAGACUUUGUUCAUCGCCUUUUU (((.(((((((((..((.((....)).))))))))))).)))........((((......)))).(((..(.((((((.....)))))))..)))........... ( -32.10) >DroSec_CAF1 7874 106 - 1 UGGCUCCUUUUGGUCCUUGAGUCCUCCAGUCGAGUGGACCCAAACAACAAGAUGUCAUAUCAUCGGAAUGGCAGUCUCAUUACGAGACUUUGUUCAUCGCCUUUUU .(((.......(((((((((.(.....).))))..)))))..........((((......)))).(((..(.((((((.....)))))))..)))...)))..... ( -28.80) >DroSim_CAF1 7863 106 - 1 UGGCUCCUUUUGGUCCUUGAGUCCUCCAAUCUAGUGGACCCAAACAACAAGAUGUCAUAUCAUCGGAAUGGCAGUCUCAUUACAAAACUUUGUUCAUCUCCUUUUU .((((((......((((((.((((...........)))).))).......((((......)))))))..)).)))).............................. ( -18.60) >DroEre_CAF1 7393 106 - 1 UGGUUGUCUUUGGUCCUCGAGUCCGCCAGUCGAGUGGACCCAAACAACAAGAUGUCAUAUCAUCGGAAUGGCAGUCUCAUUACGAGACUUUGUUCAUCGCCUUUUU ..(((((.((.(((((((((.(.....).))))..))))).)))))))..((((......)))).(((..(.((((((.....)))))))..)))........... ( -31.70) >DroYak_CAF1 8273 106 - 1 UGGUUCUUUUUGGUCCUUGAGCCCUCCAGUCGAGUGGACCCAAACAACAAGAUGUCAUAUCAUCGGAAUGGCAGUCUCAUUACGAGACUUUGUUCGUCGCCUUUUU .(((...(((.(((((((((.(......)))))..))))).)))......((((......)))).(((..(.((((((.....)))))))..)))...)))..... ( -25.50) >consensus UGGCUCCUUUUGGUCCUUGAGUCCUCCAGUCGAGUGGACCCAAACAACAAGAUGUCAUAUCAUCGGAAUGGCAGUCUCAUUACGAGACUUUGUUCAUCGCCUUUUU .(((.......(((((((((.........))))..)))))..........((((......)))).(((..(.((((((.....)))))))..)))...)))..... (-23.54 = -24.06 + 0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:26 2006