
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,070,494 – 12,070,624 |
| Length | 130 |
| Max. P | 0.859043 |

| Location | 12,070,494 – 12,070,591 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -23.89 |
| Energy contribution | -23.77 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12070494 97 + 22224390 GCCCGUUUUGCUGUAACAAAAGCACACGAAGAUGAAGUUGGAGCUGGAGC------AGGAACAGUAGGAUUCCGGA--------------CUCAGGAUCCUGGGAUCCUGGAUCCAC (((((....(((.((((...................)))).)))))).))------.((((........))))(((--------------..(((((((....)))))))..))).. ( -31.81) >DroSec_CAF1 6377 96 + 1 GCCCGUUUUGCUGUAACAAAAGCACACGAAGAUGAAGUUGGAGCUGGAGC------AGGAACAGUAGGAUUCAGGA--------------CUCAGGAUCCUGGGAUCC-GGAUCCAC .((.((((((((.((((...................)))).))).)))))------.)).......((((((.(((--------------(((((....))))).)))-)))))).. ( -30.31) >DroSim_CAF1 6359 97 + 1 GCCCGUUUUGCUGUAACAAAAGCACACGAAGAUGAAGUUGGAGCUGGAGC------AGGAACAGUAGGAUUCAGGA--------------CUCAGGAUCCUGGGAUCCUGGAUCCAC .((.((((((((.((((...................)))).))).)))))------.)).......((((((((((--------------(((((....))))).)))))))))).. ( -35.91) >DroEre_CAF1 5953 117 + 1 GCCCGUUUUGCUGUAACAAAGGCACACGAAGAUGAAGUUGGAGUGGGAGUGGGAGCAGGAGCAGUAGGAUUCAGGAUUCAGGACGCAGGACUCGGGAUCCUGGAAUCCUGGAUCCAC .(((((((..((.((((...................)))).))..))).)))).((....))....((((((((((((((((((.(.......).).)))).))))))))))))).. ( -42.31) >DroYak_CAF1 5987 104 + 1 GCCCCUUUUGCUGUAACAAAAGUACACGAAGAUUAAGUUGGGGCAGGAGU------AGGAGCAGUAGGAUUCAGGAUUCAGGA-------CUCAGUAUUCUGGGAUCCUGGAUCCAC ((((((((((((.((((...................)))).)))))))).------.)).))...........((((((((((-------(((((....))))).)))))))))).. ( -39.51) >consensus GCCCGUUUUGCUGUAACAAAAGCACACGAAGAUGAAGUUGGAGCUGGAGC______AGGAACAGUAGGAUUCAGGA______________CUCAGGAUCCUGGGAUCCUGGAUCCAC ((((.....(((.((((...................)))).))).)).))................((((((((((..............(((((....))))).)))))))))).. (-23.89 = -23.77 + -0.12)

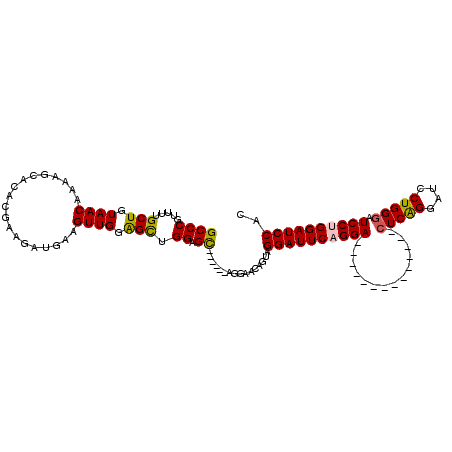
| Location | 12,070,532 – 12,070,624 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.01 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -25.40 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.787028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12070532 92 + 22224390 UGGAGCUGGAGC------AGGAACAGUAGGAUUCCGGA--------------CUCAGGAUCCUGGGAUCCUGGAUCCACGACUUUGGCAGUGGGUUGAAUGAGCUGGCGGGC ....((..((((------.((((........))))(((--------------..(((((((....)))))))..)))..).)))..)).((.((((.....)))).)).... ( -36.60) >DroSec_CAF1 6415 91 + 1 UGGAGCUGGAGC------AGGAACAGUAGGAUUCAGGA--------------CUCAGGAUCCUGGGAUCC-GGAUCCACGACUUUGGCAGUGGGUUGAAUGAGCUGGCGGGC ....((..(..(------(.........((((((.(((--------------(((((....))))).)))-)))))).((((((.......))))))..))..)..)).... ( -32.20) >DroSim_CAF1 6397 92 + 1 UGGAGCUGGAGC------AGGAACAGUAGGAUUCAGGA--------------CUCAGGAUCCUGGGAUCCUGGAUCCACGACUUUGGCAGUGGGUUGAAUGAGCUGGCGGGC ....((..(..(------(.........((((((((((--------------(((((....))))).)))))))))).((((((.......))))))..))..)..)).... ( -37.80) >DroEre_CAF1 5991 111 + 1 UGGAGUGGGAGUGGGAGCAGGAGCAGUAGGAUUCAGGAUUCAGGACGCAGGACUCGGGAUCCUGGAAUCCUGGAUCCACGACU-UGCCAGUGGGUUGAAUGAGCUGGCGGGC ........((((.(..((....))....((((((((((((((((((.(.......).).)))).))))))))))))).).)))-)(((.((.((((.....)))).)).))) ( -42.70) >DroYak_CAF1 6025 99 + 1 UGGGGCAGGAGU------AGGAGCAGUAGGAUUCAGGAUUCAGGA-------CUCAGUAUUCUGGGAUCCUGGAUCCACGACUUUGGCAGUGGGUUGAAUGAGCUGGCGGGC ....((....((------..(((..((.(((((((((((((.(((-------........))).)))))))))))))))..)))..)).((.((((.....)))).))..)) ( -35.00) >consensus UGGAGCUGGAGC______AGGAACAGUAGGAUUCAGGA______________CUCAGGAUCCUGGGAUCCUGGAUCCACGACUUUGGCAGUGGGUUGAAUGAGCUGGCGGGC ....(((((...................((((((((((..............(((((....))))).)))))))))).((((((.......))))))......))))).... (-25.40 = -26.28 + 0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:23 2006