
| Sequence ID | X_DroMel_CAF1 |
|---|---|
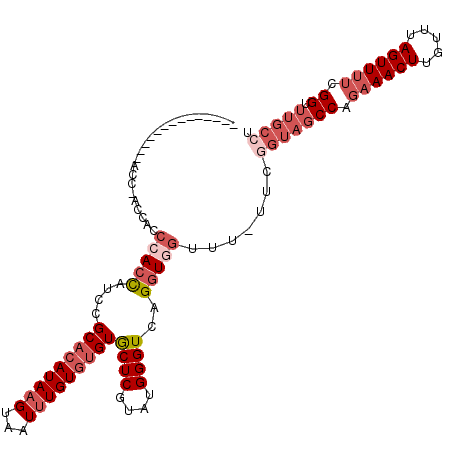
| Location | 12,067,550 – 12,067,649 |
| Length | 99 |
| Max. P | 0.954344 |

| Location | 12,067,550 – 12,067,649 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -19.61 |
| Energy contribution | -21.83 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
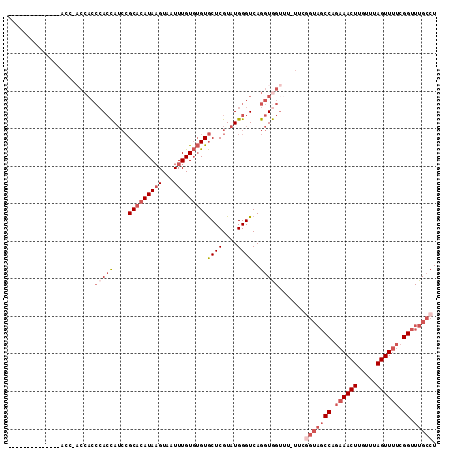
>X_DroMel_CAF1 12067550 99 + 22224390 --------------ACGAACCACCCACCAUCCGCACAUAAGUAAUUUGUGUGUGCUCGUAUGGGUCAGGUGGUUU-UUCGGUAGCCAGAAACUUGUUUAGUUUUCGGUUUGCCU --------------..((((((((..(((((((((((((((...))))))))))...).))))....))))))))-...(((((((..(((((.....)))))..)).))))). ( -35.80) >DroSec_CAF1 3430 99 + 1 --------------ACC-ACCACCCAACAUCCGCACAUAAGUAAUUUGUGUGUGCUCGUAUGGGUCAGGUGGUUUUUUUGGUAGCCAGAAACUUGUUUAGUUUUCGGUUUGCCU --------------(((-((((((((((...((((((((((...))))))))))...)).)))))..))))))......(((((((..(((((.....)))))..)).))))). ( -37.90) >DroSim_CAF1 3472 99 + 1 --------------ACC-ACCACCCACCAUCCGCACAUAAGUAAUUUGUGUGUGCUCGUAUGGGUCAGGUGGUUUUUUUGGUAGCCAGAAACUUGUUUAGUUUUCGGUUUGCCU --------------(((-((((((((.....((((((((((...))))))))))......)))))..))))))......(((((((..(((((.....)))))..)).))))). ( -36.80) >DroEre_CAF1 3288 112 + 1 GAACCACCCAUCGAACC-ACCACCCACCAUCCGCACAUAAGUAAUUUGUGUGUGCUCGUAUGGGUCAGGUGGUUU-UUCGGUAGCCAGAAACUUGUUUAGUUUUCGGUUUGCCU ............(((((-((((((((.....((((((((((...))))))))))......)))))..))))))))-...(((((((..(((((.....)))))..)).))))). ( -39.60) >DroYak_CAF1 3049 88 + 1 -------------------------ACCAUCCGCACAUAAGUAAUUUGUGUGUGCUCGUAUGGGUCAGGUGGUUU-UUCAGUAGCCAAAAACUUGUUUAGUUAACGGUUUGCCU -------------------------.(((((((((((((((...))))))))))...).))))...((((((((.-......))))..(((((.(((.....)))))))))))) ( -20.70) >DroAna_CAF1 3278 81 + 1 -------------------CCACCCACUAG--GCGUAUACGUAAUUUGU--GUACUCGUGUGGGUCAGGUAGCC----------CCAGAAACUUGUUUAGUUUUCGGUUUGGCU -------------------((((((((.(.--(.(((((((.....)))--)))).).)))))))..)).((((----------((..(((((.....)))))..))...)))) ( -28.30) >consensus ______________ACC_ACCACCCACCAUCCGCACAUAAGUAAUUUGUGUGUGCUCGUAUGGGUCAGGUGGUUU_UUCGGUAGCCAGAAACUUGUUUAGUUUUCGGUUUGCCU .......................(((((....(((((((((...)))))))))((((....))))..))))).......(((((((.((((((.....)))))).)).))))). (-19.61 = -21.83 + 2.22)

| Location | 12,067,550 – 12,067,649 |
|---|---|
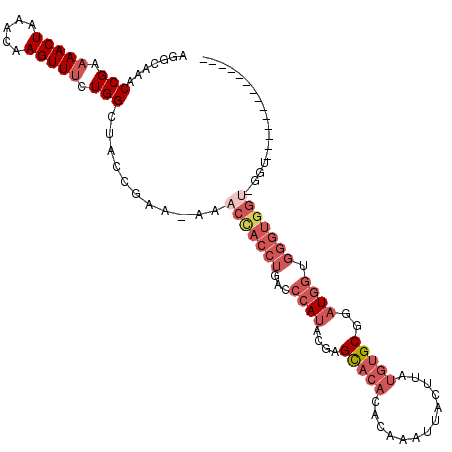
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -15.34 |
| Energy contribution | -17.59 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12067550 99 - 22224390 AGGCAAACCGAAAACUAAACAAGUUUCUGGCUACCGAA-AAACCACCUGACCCAUACGAGCACACACAAAUUACUUAUGUGCGGAUGGUGGGUGGUUCGU-------------- .((....(((.(((((.....))))).)))...))...-.((((((((...((((....(((((.............)))))..)))).))))))))...-------------- ( -27.22) >DroSec_CAF1 3430 99 - 1 AGGCAAACCGAAAACUAAACAAGUUUCUGGCUACCAAAAAAACCACCUGACCCAUACGAGCACACACAAAUUACUUAUGUGCGGAUGUUGGGUGGU-GGU-------------- .((....(((.(((((.....))))).)))...))......((((((..(((((.(((.(((((.............)))))...)))))))))))-)))-------------- ( -28.52) >DroSim_CAF1 3472 99 - 1 AGGCAAACCGAAAACUAAACAAGUUUCUGGCUACCAAAAAAACCACCUGACCCAUACGAGCACACACAAAUUACUUAUGUGCGGAUGGUGGGUGGU-GGU-------------- .((....(((.(((((.....))))).)))...))......((((((..((((((....(((((.............))))).....)))))))))-)))-------------- ( -30.12) >DroEre_CAF1 3288 112 - 1 AGGCAAACCGAAAACUAAACAAGUUUCUGGCUACCGAA-AAACCACCUGACCCAUACGAGCACACACAAAUUACUUAUGUGCGGAUGGUGGGUGGU-GGUUCGAUGGGUGGUUC .......(((.(((((.....))))).)))(((((.(.-.(((((((..((((((....(((((.............))))).....)))))))))-))))...).)))))... ( -36.32) >DroYak_CAF1 3049 88 - 1 AGGCAAACCGUUAACUAAACAAGUUUUUGGCUACUGAA-AAACCACCUGACCCAUACGAGCACACACAAAUUACUUAUGUGCGGAUGGU------------------------- (((....(((..((((.....))))..)))....((..-....)))))...((((....(((((.............)))))..)))).------------------------- ( -14.12) >DroAna_CAF1 3278 81 - 1 AGCCAAACCGAAAACUAAACAAGUUUCUGG----------GGCUACCUGACCCACACGAGUAC--ACAAAUUACGUAUACGC--CUAGUGGGUGG------------------- ((((...(((.(((((.....))))).)))----------)))).....((((((.((.((((--.........)))).)).--...))))))..------------------- ( -21.50) >consensus AGGCAAACCGAAAACUAAACAAGUUUCUGGCUACCGAA_AAACCACCUGACCCAUACGAGCACACACAAAUUACUUAUGUGCGGAUGGUGGGUGGU_GGU______________ .......(((.(((((.....))))).)))...........(((((((...((((....(((((.............)))))..)))).))))))).................. (-15.34 = -17.59 + 2.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:20 2006