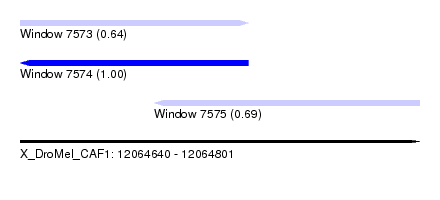
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,064,640 – 12,064,801 |
| Length | 161 |
| Max. P | 0.999022 |

| Location | 12,064,640 – 12,064,732 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 91.96 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12064640 92 + 22224390 UAUUCGUCUUAAUUUUCUGCAAGUGCAUUCACCUGCACACCUGUGCAAAUAAUUGUAAUUAAGUCGAGUGUCAUCGCGAAUUUACAACGCCA ((((((.((((((....((((((((....))).(((((....))))).....))))))))))).)))))).....(((.........))).. ( -21.00) >DroSec_CAF1 473 92 + 1 UAUUCGACUUAAUUUUCUGCAAGUGCAUUCUCCUGCACACCUGUGCAAAUAAUUGUAAUUAAGUCGAGUGUCAUCGCGAAUUUACAACGCCA ((((((((((((((...(((....)))......(((((....))))).........)))))))))))))).....(((.........))).. ( -24.80) >DroSim_CAF1 520 92 + 1 UAUUCGACUUAAUUUUCUGCAAGUGCAUUCACCUGCACACCUGUGCAAAUAAUUGUAAUUAAGUCGAGUGUCAUCGCGAAUUUACAACGCCA (((((((((((((....((((((((....))).(((((....))))).....)))))))))))))))))).....(((.........))).. ( -24.90) >DroEre_CAF1 424 92 + 1 AAUUGCACCUAUUUUUCCGCAAGUGCAGUCACCUGCACACCUGUGCAAAUAAUUGUAAUUAAGUCGAGUGUCAUCGCGAAUUUACAACGCCA ..((((((.........(....)(((((....))))).....))))))....((((((....(.(((......))))....))))))..... ( -19.90) >DroYak_CAF1 318 82 + 1 ---------UA-UUUUCUGCAAGUGCAUUCACCUGCACACCUGUGCAAAUAAUUGUAAUUAAGUCGAGUGUCAUCGCGAGUUUACAACGCCA ---------..-.....((((.(((((......))))).....)))).....((((((....(.(((......))))....))))))..... ( -15.70) >consensus UAUUCGACUUAAUUUUCUGCAAGUGCAUUCACCUGCACACCUGUGCAAAUAAUUGUAAUUAAGUCGAGUGUCAUCGCGAAUUUACAACGCCA ......................(.((.......(((((....))))).....((((((....(.(((......))))....)))))).))). (-15.20 = -15.20 + 0.00)



| Location | 12,064,640 – 12,064,732 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.96 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -22.48 |
| Energy contribution | -24.60 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12064640 92 - 22224390 UGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUACAAUUAUUUGCACAGGUGUGCAGGUGAAUGCACUUGCAGAAAAUUAAGACGAAUA ((.((((((......)))))).)).(((.(((((((....((((((((((....)))))))))).(((....)))...))))))).)))... ( -25.80) >DroSec_CAF1 473 92 - 1 UGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUACAAUUAUUUGCACAGGUGUGCAGGAGAAUGCACUUGCAGAAAAUUAAGUCGAAUA ((.((((((......)))))).)).(((((((((((.......(((((((....)))))))....(((....)))...)))))))))))... ( -27.50) >DroSim_CAF1 520 92 - 1 UGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUACAAUUAUUUGCACAGGUGUGCAGGUGAAUGCACUUGCAGAAAAUUAAGUCGAAUA ((.((((((......)))))).)).(((((((((((....((((((((((....)))))))))).(((....)))...)))))))))))... ( -31.20) >DroEre_CAF1 424 92 - 1 UGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUACAAUUAUUUGCACAGGUGUGCAGGUGACUGCACUUGCGGAAAAAUAGGUGCAAUU ..(((((((((.....(((......)))......)))))))..((((((.....((((((....)))))).)))))).........)).... ( -24.90) >DroYak_CAF1 318 82 - 1 UGGCGUUGUAAACUCGCGAUGACACUCGACUUAAUUACAAUUAUUUGCACAGGUGUGCAGGUGAAUGCACUUGCAGAAAA-UA--------- ..(((((((((.....(((......)))......))))).((((((((((....))))))))))))))............-..--------- ( -18.60) >consensus UGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUACAAUUAUUUGCACAGGUGUGCAGGUGAAUGCACUUGCAGAAAAUUAAGUCGAAUA ((.((((((......)))))).)).(((((((((((....((((((((((....)))))))))).(((....)))...)))))))))))... (-22.48 = -24.60 + 2.12)

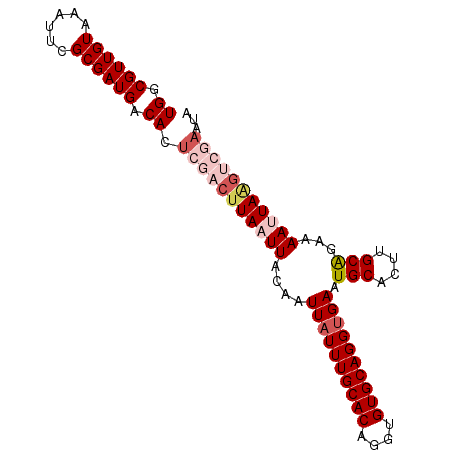
| Location | 12,064,694 – 12,064,801 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.56 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -27.76 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12064694 107 - 22224390 CCAUGCGAUACGGUUGACCCGAGUUGACCUGUGCAUAAUCACCAU-UUGCCUUUGUUCCCGGCAUUUUGAUGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUAC .(((((((...(((..((....))..)))..((((....(.((((-(((((.........))))....))))).).))))...)))).)))................. ( -25.20) >DroSec_CAF1 527 108 - 1 CAAUGCGAUACGGUUGACCCGAGUUGACCUGUGCAUAAUCGCCAUUUUGCCUUUGUUCCCGGCAUUUUGAUGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUAC ((.(((((...(((..((....))..)))..((((....(((((((.((((.........))))....))))))).))))...))))).))................. ( -31.90) >DroSim_CAF1 574 108 - 1 CAAUGCGAUACGGUUGACCCGAGUUGACCUGUGCAUAAUCGCCAUUUUGCCUUUGUUCCCGGCAUUUUGAUGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUAC ((.(((((...(((..((....))..)))..((((....(((((((.((((.........))))....))))))).))))...))))).))................. ( -31.90) >DroEre_CAF1 478 107 - 1 CACUGCGAUUCGGUUGACCCGAGUUGACCUGUGCAUAAUCGCCAU-UUGCCUUUGUGCCCGGCAUUUUGAUGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUAC ((.(((((...(((..((....))..)))..((((....((((((-(((((.........))))....))))))).))))...))))).))................. ( -30.60) >DroYak_CAF1 362 107 - 1 CAAUGCGAUACGGUUGAUCCGAGUUGACCUGUGCAUAAUCGCCAU-UUGCCUUUGUCCCCGGCAUUUUGAUGGCGUUGUAAACUCGCGAUGACACUCGACUUAAUUAC ((.(((((...(((..((....))..)))..((((....((((((-(((((.........))))....))))))).))))...))))).))................. ( -28.90) >consensus CAAUGCGAUACGGUUGACCCGAGUUGACCUGUGCAUAAUCGCCAU_UUGCCUUUGUUCCCGGCAUUUUGAUGGCGUUGUAAAUUCGCGAUGACACUCGACUUAAUUAC ((.(((((...(((..((....))..)))..((((....((((((..((((.........)))).....)))))).))))...))))).))................. (-27.76 = -28.00 + 0.24)

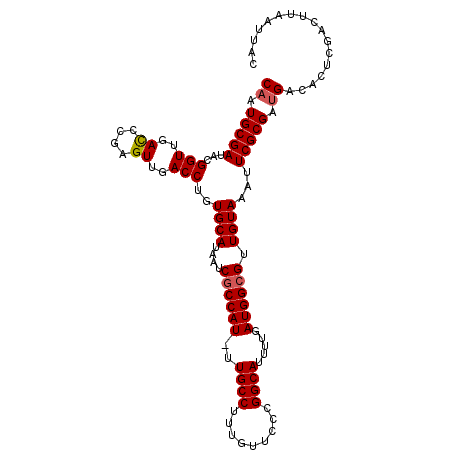

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:17 2006