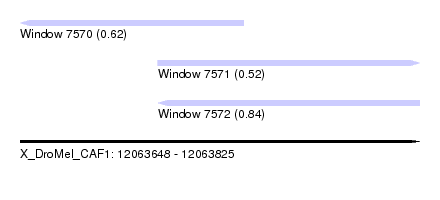
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,063,648 – 12,063,825 |
| Length | 177 |
| Max. P | 0.840859 |

| Location | 12,063,648 – 12,063,747 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -12.85 |
| Energy contribution | -13.13 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12063648 99 - 22224390 AUAUUGUCCGCGAUUCAAUGAAAAUCUCUGC--GUCGAAAUCCCUAUCUAUCGGAUUUCCAAUCGCUC-UGAAUCGCCGGUUAUCUUUUAU-CCU-UAAAAUGU---------------- .......((((((((((..((.....)).((--(..(((((((.........)))))))....)))..-))))))).)))...........-...-........---------------- ( -21.80) >DroSec_CAF1 95101 98 - 1 AUAUUCCCAGCGAUUCAAUGAAAAUCUCUGC--GUCGAAAUCCCUAUCUAUCGGAUUUCCAAUCGCUC-UGGAUCGCCGGUUAUCU-GUAU-CCU-UAAAGUGU---------------- ......((.((((((((..((.....)).((--(..(((((((.........)))))))....)))..-)))))))).))......-....-...-........---------------- ( -22.50) >DroSim_CAF1 72156 98 - 1 AUAUUCCCUGUGAUUCAAUGAAAAUCUCUGC--GUCGAAAUCCCUAUCUAUCGGAUUUCCAAUCGCUC-UGGAUCGCCGGUUAUCU-GUAU-CCU-UAAAGUGU---------------- ......((.((((((((..((.....)).((--(..(((((((.........)))))))....)))..-)))))))).))......-....-...-........---------------- ( -20.60) >DroEre_CAF1 92800 97 - 1 GUAUUCCCCGCGAUUCAAUGAAAAUCUCUGC--GUCGAAAUCCGUAUCUGUCGGAUUUCCACUCGCUC-UGGAUCGCCGGUAAUCU-UU-U-CCU-UAAAGUGU---------------- ......((.((((((((..((.....)).((--(..((((((((.......))))))))....)))..-)))))))).))......-..-.-...-........---------------- ( -26.00) >DroYak_CAF1 95056 98 - 1 AUAUUCACCGUGAUUCAAUGAAAAUCUCUGC--GUCGAAAUCCCUAUCUAUCGGAUUUUCACUCGCUC-UGGAUCGCCGUUUAGCU-UU-U-CCUUUAAAGUGU---------------- ......((.((((((((..((.....)).((--(..(((((((.........)))))))....)))..-)))))))).))...(((-((-.-.....)))))..---------------- ( -19.00) >DroAna_CAF1 35363 111 - 1 GUCA-GCCUUCGAUCGAAUGGAAAUCCUUUCCGGGCGAACUCCU-----CUCCGCUUCC--UUGGCUUGGGGAUCUCUUGUUUUUUUGUAAAUAU-UAUAGUUUUAUGUUUUGUUAUCGA ....-....(((((((..(((((.....)))))..)))..((((-----(...(((...--..)))..))))).................((((.-.(((......)))..)))).)))) ( -20.60) >consensus AUAUUCCCCGCGAUUCAAUGAAAAUCUCUGC__GUCGAAAUCCCUAUCUAUCGGAUUUCCAAUCGCUC_UGGAUCGCCGGUUAUCU_GUAU_CCU_UAAAGUGU________________ ......((.((((((((..((.....)).((.....(((((((.........))))))).....))...)))))))).))........................................ (-12.85 = -13.13 + 0.28)

| Location | 12,063,709 – 12,063,825 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -26.39 |
| Energy contribution | -28.28 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.520972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12063709 116 + 22224390 UUUCGAC--GCAGAGAUUUUCAUUGAAUCGCGGACAAUAUAUAUCCAUACGCGGGUGGCAA--UGCCACCGAGGCCCCUGUGGGCCCCUCCUGCCCCUGCCCAUGCAUCGAAACGGGGUG ..((..(--((.(.(((((.....))))).)(((.........)))....)))((((((..--.))))))))(((((....)))))......((((((((....))).......))))). ( -37.81) >DroSec_CAF1 95161 107 + 1 UUUCGAC--GCAGAGAUUUUCAUUGAAUCGCUGGGAAUAUAUAUCCAUACGCGGGUGGCAA--UGCCACCGAGGCCCCUGUGGGCCCCUCCUGCCCCUGCCCAUGCAUCGA--------- ..((((.--(((..(((((.....)))))((.(((..................((((((..--.))))))(.(((((....))))).)......))).))...))).))))--------- ( -37.60) >DroSim_CAF1 72216 107 + 1 UUUCGAC--GCAGAGAUUUUCAUUGAAUCACAGGGAAUAUAUAUCCAUACGCGGGUGGCAA--UGCCACCGAGGCCCCUGUGGGCCCCUCCUGCCCCUGCCCAUGCAUCGA--------- ..((((.--(((..(((((.....))))).(((((..................((((((..--.))))))(.(((((....))))).)......)))))....))).))))--------- ( -38.60) >DroEre_CAF1 92859 107 + 1 UUUCGAC--GCAGAGAUUUUCAUUGAAUCGCGGGGAAUACACAUCCAUCCGCGGGUGGCAA--UGCCACCGAGGCCCCUGUGGGCCCCUCCAGCCCCUGUCCAUGCAUCGA--------- ..((((.--(((..(((.....(((...((((((((.......)))..)))))((((((..--.))))))(.(((((....))))).)..))).....)))..))).))))--------- ( -40.50) >DroYak_CAF1 95116 107 + 1 UUUCGAC--GCAGAGAUUUUCAUUGAAUCACGGUGAAUAUAUAUCCAUCUGCGGGUGGCAA--UGCCACCGAGGCCCCUGUGGGCCCCUCCUGCCCCUGCCCAUGCAUCGA--------- ..(((((--((((((((.(((((((.....))))))).....)))..))))))((((((..--.))))))(.(((((....))))).).........(((....)))))))--------- ( -37.30) >DroAna_CAF1 35435 110 + 1 GUUCGCCCGGAAAGGAUUUCCAUUCGAUCGAAGGC-UGACAUAUCCUGCUGCGGGUGGCGAUAUGCCACCAAGGCCCCCUCCGGGCCGUCCUGCCCCUGGCCGCGACCUCA--------- ..((((..((..(((.......(((....)))(((-.(((.............((((((.....))))))..(((((.....))))))))..))))))..)))))).....--------- ( -42.10) >consensus UUUCGAC__GCAGAGAUUUUCAUUGAAUCGCGGGGAAUAUAUAUCCAUACGCGGGUGGCAA__UGCCACCGAGGCCCCUGUGGGCCCCUCCUGCCCCUGCCCAUGCAUCGA_________ ..((((...(((..(((((.....))))).(((((..................((((((.....))))))..(((((....)))))........)))))....))).))))......... (-26.39 = -28.28 + 1.89)


| Location | 12,063,709 – 12,063,825 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -46.96 |
| Consensus MFE | -33.46 |
| Energy contribution | -35.04 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12063709 116 - 22224390 CACCCCGUUUCGAUGCAUGGGCAGGGGCAGGAGGGGCCCACAGGGGCCUCGGUGGCA--UUGCCACCCGCGUAUGGAUAUAUAUUGUCCGCGAUUCAAUGAAAAUCUCUGC--GUCGAAA .......((((((((((.((((....((....(((((((....)))))))((((((.--..)))))).))....(((((.....)))))))((((.......)))))))))--))))))) ( -47.40) >DroSec_CAF1 95161 107 - 1 ---------UCGAUGCAUGGGCAGGGGCAGGAGGGGCCCACAGGGGCCUCGGUGGCA--UUGCCACCCGCGUAUGGAUAUAUAUUCCCAGCGAUUCAAUGAAAAUCUCUGC--GUCGAAA ---------((((((((.((((.(((((....(((((((....)))))))((((((.--..)))))).))(((((....))))).))).))((((.......)))))))))--))))).. ( -45.80) >DroSim_CAF1 72216 107 - 1 ---------UCGAUGCAUGGGCAGGGGCAGGAGGGGCCCACAGGGGCCUCGGUGGCA--UUGCCACCCGCGUAUGGAUAUAUAUUCCCUGUGAUUCAAUGAAAAUCUCUGC--GUCGAAA ---------((((((((.((((((((((....(((((((....)))))))((((((.--..)))))).))(((((....))))).))))))((((.......)))))))))--))))).. ( -48.80) >DroEre_CAF1 92859 107 - 1 ---------UCGAUGCAUGGACAGGGGCUGGAGGGGCCCACAGGGGCCUCGGUGGCA--UUGCCACCCGCGGAUGGAUGUGUAUUCCCCGCGAUUCAAUGAAAAUCUCUGC--GUCGAAA ---------((((((((.(((.......(((((((((((....)))))))((((((.--..))))))(((((..(((.......)))))))).)))).......))).)))--))))).. ( -50.14) >DroYak_CAF1 95116 107 - 1 ---------UCGAUGCAUGGGCAGGGGCAGGAGGGGCCCACAGGGGCCUCGGUGGCA--UUGCCACCCGCAGAUGGAUAUAUAUUCACCGUGAUUCAAUGAAAAUCUCUGC--GUCGAAA ---------((((((((((((((.((((....(((((((....)))))))((((((.--..)))))).))...(((((....))))))).)).))))..((.....)))))--))))).. ( -44.90) >DroAna_CAF1 35435 110 - 1 ---------UGAGGUCGCGGCCAGGGGCAGGACGGCCCGGAGGGGGCCUUGGUGGCAUAUCGCCACCCGCAGCAGGAUAUGUCA-GCCUUCGAUCGAAUGGAAAUCCUUUCCGGGCGAAC ---------....(((.(.(((...))).)))).(((((((((((((...((((((.....)))))).))..............-.(((((....))).))....))))))))))).... ( -44.70) >consensus _________UCGAUGCAUGGGCAGGGGCAGGAGGGGCCCACAGGGGCCUCGGUGGCA__UUGCCACCCGCGGAUGGAUAUAUAUUCCCCGCGAUUCAAUGAAAAUCUCUGC__GUCGAAA .........(((((......((((((((....(((((((....)))))))((((((.....)))))).))...(((((..............)))))........))))))..))))).. (-33.46 = -35.04 + 1.58)

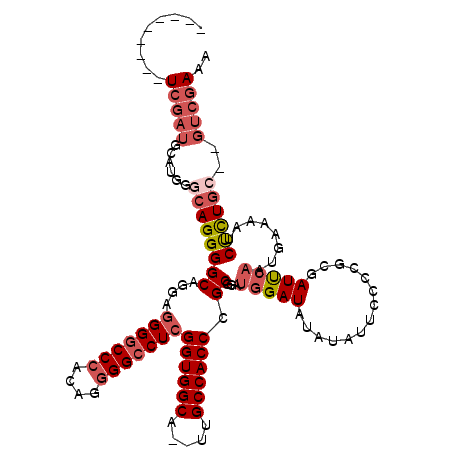
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:15 2006