
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,063,481 – 12,063,580 |
| Length | 99 |
| Max. P | 0.554037 |

| Location | 12,063,481 – 12,063,580 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.84 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -9.88 |
| Energy contribution | -10.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12063481 99 - 22224390 UUGGUAAG-----------UAAUUUGAGUGCUGUUGUC-UAGAAGACAACAACCUUGUGACCUUUCGUUACGCUUUACUAAAUCCUUUUCGUGCUUCUCUUUUAUCAAGUC--------- ((((((((-----------......(((..((((((((-.....))))))).....(((((.....)))))...................)..)))....))))))))...--------- ( -22.10) >DroSec_CAF1 94940 98 - 1 UGGGUGAG-----------UAAU-UGAGUAGGGUUGUC-UUAAAGACAACAACCUUGUGACCUUUCGUUACGCUUUACUAAAUCCAUUUUGUGUUUCUCUUUUAUCAAGUU--------- (((((.((-----------((..-.(((((((((((((-.....))))))..))).(((((.....)))))))))))))..))))).........................--------- ( -20.40) >DroSim_CAF1 71995 98 - 1 UGGGUGAG-----------UAAU-UGAGUGGGGUUGUC-UUAAAGACAACAACCUUGUGACCUUUCGUUACGCUUUACUAAAUCCAUUUUGUAUUUCUCUUUUAUCAAGUU--------- .(((.(((-----------((..-.(((((((((((((-.....))))))......(((((.....)))))...........)))))))..))))))))............--------- ( -22.70) >DroEre_CAF1 92631 106 - 1 UAUGUGAG-----------UAAU-UAAGUGGGGUUGUC-UUAAAGACAACAACCUUGUGACCU-UCGUUACGCUUUACUAUGUCCUUUUCCUGUUUCUCUUUGAUCAGUUAAUUUCUCUU .....(((-----------.(((-(((.(((.((((((-.....))))))......(((((..-..))))).................................))).)))))).))).. ( -21.00) >DroYak_CAF1 94885 107 - 1 UAUGUGAG-----------UAAU-UUAGUGGGGUUGUC-UUGAAGACAACAACCUUGUGACCUUUUGUUACGCUUUACUAAGUCCUUUUUCUGUUUCACUUCCAUCAAUUAUAUACACUU ..((((.(-----------((((-(..(((((((((((-.....))))))......(((((.....)))))((((....)))).................))))).)))))).))))... ( -21.10) >DroAna_CAF1 35209 98 - 1 UGGGGGAAAGAGGUUUAAAAAGU-UAAUGGAAGUUGCGUUUUGAGACAACAACCUUGUGACUUUUCGUUAUGCUUUACUUUCUUUACUU-----------UUUAUCC-UUU--------- ........(((((..((((((((-....((((((.((((..(((((..(((....)))....)))))..))))...))))))...))))-----------)))).))-)))--------- ( -25.10) >consensus UGGGUGAG___________UAAU_UGAGUGGGGUUGUC_UUAAAGACAACAACCUUGUGACCUUUCGUUACGCUUUACUAAAUCCAUUUUGUGUUUCUCUUUUAUCAAGUU_________ ..........................((((((((((((......))))))......(((((.....)))))..))))))......................................... ( -9.88 = -10.38 + 0.50)

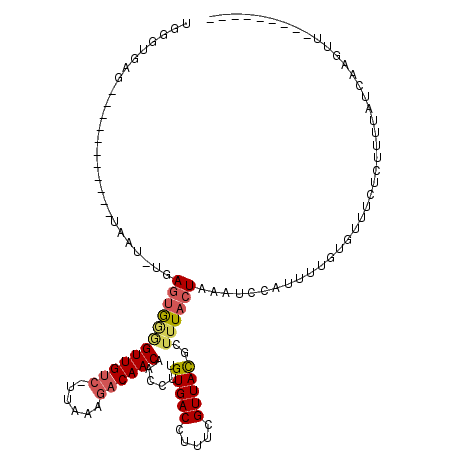
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:12 2006