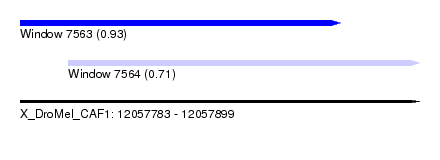
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,057,783 – 12,057,899 |
| Length | 116 |
| Max. P | 0.926107 |

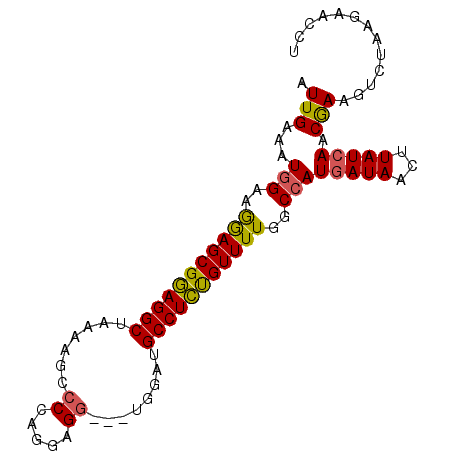
| Location | 12,057,783 – 12,057,876 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -21.26 |
| Energy contribution | -20.89 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12057783 93 + 22224390 AUUGAAAUGGAAGGAGCGUAGGCUAAAAGCCCCAGGAGGUGGUGGAUGCCUAUGUUUUGGCAAUGAUAACUUAUCAACGAAGUCUAAGAACCU .......((((.(((((((((((.....(((((....)).)))....))))))))))).....(((((...)))))......))))....... ( -24.30) >DroSec_CAF1 89373 90 + 1 AUUGAAUUGGUAGGAGCGGAGGCUAAAAGCCCGAGGAGG---UGGAUGCCUCUGUUUUGGCCAUGAUAACUUAUCAACGAAGUCUAAGAACCU .(((...((((..((((((((((.....(((......))---)....))))))))))..))))(((((...))))).)))............. ( -27.80) >DroEre_CAF1 87045 90 + 1 AUUGAAAUGGUUAGAGCGGAGGCUAAAAGCCCCGGGAGU---UGGAUGCCUCUGUUUUGGCCAUGAUAACUUAUCAACAAAGUCUAAGAACCU .(((..(((((((((((((((((....((((....).))---)....)))))))))))))))))((((...))))..)))............. ( -30.30) >DroYak_CAF1 89268 88 + 1 AUUGAAAUGGAAGAAGCGGAGGCUAAAAGCCCCUGGAGG---UGGAUGCCUCCGUUUUGGCCAUAAUAACUUAUCAACGAAGUCUA--AACCU ......((((..(((((((((((......((((....))---.))..)))))))))))..))))......................--..... ( -29.20) >consensus AUUGAAAUGGAAGGAGCGGAGGCUAAAAGCCCCAGGAGG___UGGAUGCCUCUGUUUUGGCCAUGAUAACUUAUCAACGAAGUCUAAGAACCU .(((...(((..(((((((((((.......((.....))........)))))))))))..)))(((((...))))).)))............. (-21.26 = -20.89 + -0.38)

| Location | 12,057,797 – 12,057,899 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 89.77 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.21 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12057797 102 + 22224390 AGCGUAGGCUAAAAGCCCCAGGAGGUGGUGGAUGCCUAUGUUUUGGCAAUGAUAACUUAUCAACGAAGUCUAAGAACCUGGCUUCUGACAUUCCAAGAAAA-A .((((((((.....(((((....)).)))....))))))))(((((.((((........(((..((((((.........))))))))))))))))))....-. ( -29.10) >DroSec_CAF1 89387 99 + 1 AGCGGAGGCUAAAAGCCCGAGGAGG---UGGAUGCCUCUGUUUUGGCCAUGAUAACUUAUCAACGAAGUCUAAGAACCUGGCUUCUGACAUGCCAAGAAAA-A .((((((((.....(((......))---)....))))))))((((((..(((((...)))))..((((((.........))))))......))))))....-. ( -31.50) >DroEre_CAF1 87059 98 + 1 AGCGGAGGCUAAAAGCCCCGGGAGU---UGGAUGCCUCUGUUUUGGCCAUGAUAACUUAUCAACAAAGUCUAAGAACCUGGCUUCUGACAUGCCAAGAAAA-- .((((((((....((((....).))---)....))))))))((((((..(((((...))))).(((((((.........))))).))....))))))....-- ( -27.50) >DroYak_CAF1 89282 98 + 1 AGCGGAGGCUAAAAGCCCCUGGAGG---UGGAUGCCUCCGUUUUGGCCAUAAUAACUUAUCAACGAAGUCUA--AACCUGGCUUCUGACAUGAAAACAAAAAA ......((((((((.....((((((---(....)))))))))))))))................((((((..--.....)))))).................. ( -26.50) >consensus AGCGGAGGCUAAAAGCCCCAGGAGG___UGGAUGCCUCUGUUUUGGCCAUGAUAACUUAUCAACGAAGUCUAAGAACCUGGCUUCUGACAUGCCAAGAAAA_A (((((((((.......((.....))........)))))))))((((.((((........(((..((((((.........)))))))))))))))))....... (-20.90 = -21.21 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:08 2006