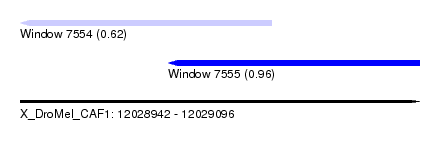
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,028,942 – 12,029,096 |
| Length | 154 |
| Max. P | 0.960238 |

| Location | 12,028,942 – 12,029,039 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.24 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12028942 97 - 22224390 GUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAUUGAAUUGCACACUGC-------UGACGUUCGCGGUGCUGCCAUCGCAGUGCUGUCAAAGUACAG (((((((((....)))))(((((((..((......))....((...(((((((((-------........)))))).)))..))))))))).......)))).. ( -35.50) >DroSec_CAF1 61156 97 - 1 GUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAUUGAAUUGCACACUGC-------UGACGUUCGCGGUGCUGCCAUCGCAGUGCUGUCAAAGUACAG (((((((((....)))))(((((((..((......))....((...(((((((((-------........)))))).)))..))))))))).......)))).. ( -35.50) >DroSim_CAF1 59756 97 - 1 GUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAUUGAAUUGCACACUGC-------UGACGUUCGCGGUGCUGCCAUCGCAGUGCUGUCAAAGUACAG (((((((((....)))))(((((((..((......))....((...(((((((((-------........)))))).)))..))))))))).......)))).. ( -35.50) >DroEre_CAF1 58817 104 - 1 GUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAUUGAAUUGCACACUGCUGACUGCUGACGGUCGCGGUGCUGCCAUCGCAGUGCUGUCCAAGUACAG (((((((((....)))))(((((((..((......))....((...(((((((((.(((((....))))))))))).)))..))))))))).......)))).. ( -41.90) >DroYak_CAF1 60891 97 - 1 GUAUCGGCCAUUGGGCCGGCGCUGCCAACUUGAUUGUAACUGAAUUGCACACUGC-------UGACGUUCGCGGUGCUGCCAUCGCAGUGCUGUCAAAGUACAG (((((((((....)))))(((((((..((......))....((...(((((((((-------........)))))).)))..))))))))).......)))).. ( -35.40) >DroAna_CAF1 1965 90 - 1 GUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAUUGAAUUGCACACUGC-------UGACGUUCGCGGAGCUGCUAUU-CGGUG------GGGUACAG (((((((((....))))....(..((.((......))....((((.((((.((((-------........)))).).))).)))-)))..------)))))).. ( -30.20) >consensus GUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAUUGAAUUGCACACUGC_______UGACGUUCGCGGUGCUGCCAUCGCAGUGCUGUCAAAGUACAG (((((((((....)))))(((((((..((......)).........(((((((((...............)))))).)))....))))))).......)))).. (-29.88 = -30.24 + 0.36)


| Location | 12,028,999 – 12,029,096 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.76 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12028999 97 - 22224390 GCGGAAAAGCGGUGGGCGGU-GGA----------AUGGGUGGUGUAAUAUCAGAGAGGAUGUACCAUUGUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAU ((......))(((.((((((-(..----------...((((((...(((((......)))))))))))...((((((....))))))))))))).))).......... ( -33.90) >DroSec_CAF1 61213 107 - 1 GCGGAAAAGCGGUGGGCGGU-GGGUGGUCGGGCUAUGGGUGGUGUAAUAUCAGAGAGGAUGUACCAUUGUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAU ..((...((((.(((.(...-.(((((((((......((((((...(((((......)))))))))))...))))))))).).))).)))).)).............. ( -36.20) >DroSim_CAF1 59813 100 - 1 GCGGAAAAGCGGUGGGCGGU-GGGCGGUGA-------GGUGGUGUAAUAUCAGAGAGGAUGUACCAUUGUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAU ..((...((((.(((.((((-((.(((((.-------.(((((...(((((......))))))))))..))))).))))).).))).)))).)).............. ( -39.30) >DroEre_CAF1 58881 93 - 1 GCGGAAAAGCGGUGGGCGGGU---------------GGCUGGUAUAAUGUCAGAGAGGAUGUACCAUUGUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAU ..((...((((.(((.(..((---------------(((((((((((((.((.......))...)))))))))))))))..).))).)))).)).............. ( -39.50) >DroYak_CAF1 60948 101 - 1 GCGGAAAAGCGCUGGGGGGUUGGGUGAU-------UGGGUGGUAUAAUAUCAGAGAGGAUGUACCAUUGUAUCGGCCAUUGGGCCGGCGCUGCCAACUUGAUUGUAAC ..((...((((((((.....(((.((((-------..(((((((((...((...))...)))))))))..)))).))).....)))))))).)).............. ( -36.30) >consensus GCGGAAAAGCGGUGGGCGGU_GGG_G_U_______UGGGUGGUGUAAUAUCAGAGAGGAUGUACCAUUGUAUCGGCCAUUAGGCCGGCGCUGCCAACUUGAUUGUAAU ((......))(((.((((((.................((((((...(((((......)))))))))))...((((((....)))))).)))))).))).......... (-27.12 = -27.76 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:00 2006