
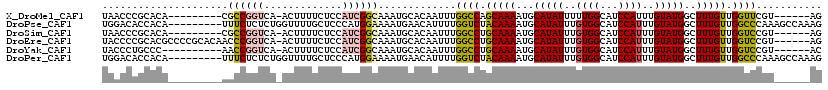
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,025,862 – 12,025,966 |
| Length | 104 |
| Max. P | 0.582677 |

| Location | 12,025,862 – 12,025,966 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
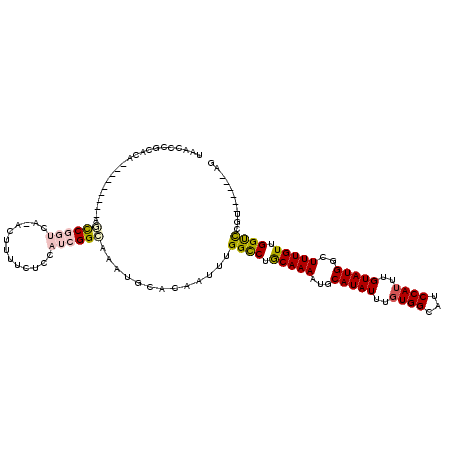
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12025862 104 - 22224390 UAACCCGCACA---------CGCCGGUCA-ACUUUUCUCCAUCGGCAAAUGCACAAUUUGGCCAGCAAAAUGCAUAUUUUUGGCAUCCAUUUGUAUGGCUUUGUUGGUUCGU------AG ......(((..---------.((((((..-..........))))))...))).((((..((((((((((((((.........))))...))))).)))))..))))......------.. ( -26.80) >DroPse_CAF1 157699 111 - 1 UGGACACCACA---------UUUCUCUCUGGUUUUGCUCCCAUGGAAAAUGAACAUUUUGGUCUACAAAAUGCAUAUUUGUGGCAUCCAUUUGUAUGGCUUUGUUGGCCCAAAGCCAAAG ((((..((((.---------....(((.(((........))).)))(((((..(((((((.....)))))))..)))))))))..))))......((((((((......))))))))... ( -33.30) >DroSim_CAF1 56779 104 - 1 UAACCCGCACA---------CGCCGGUCA-ACUUUUCUCCAUCGGCAAAUGCACAAUUUGGCCUGCAAAAUGCAUAUUUGUGGCAUCCAUUUGUAUGGCUUUGUUGGUCCGU------AG ......(((..---------.((((((..-..........))))))...))).((((..((((((((((((((.........))))...)))))).))))..))))......------.. ( -27.20) >DroEre_CAF1 54441 113 - 1 UACCCCGCACGCCCCGCACAACCCGGUCA-ACUUUUCUCCAUCGGCAAAUGCACAAUUUGGCCUGCAAAAUGCAUAUUUGUGGCAUCCAUUUGUAUGGCUUUGUUGGUCCGU------AG ........(((..(((((....(((((..-..........)))))....)))..(((..((((((((((((((.........))))...)))))).))))..)))))..)))------.. ( -25.60) >DroYak_CAF1 57802 103 - 1 UACCCUGCCC----------AACCGGUCA-ACUUUUCUCCAUCGGCAAAUGCACAAUUUGGCCUGCAAAAUGCAUAUUUGUGGCAUCCAUUUGUAUGGCUUUGUUGGUCCGU------AC ..........----------...(((.(.-..............((....)).((((..((((((((((((((.........))))...)))))).))))..))))).))).------.. ( -22.70) >DroPer_CAF1 162827 111 - 1 UGGACACCACA---------UUUCUCUCUGGUUUUGCUCCCAUGGAAAAUGAACAUUUUGGUCUACAAAAUGCAUAUUUGUGGCAUCCAUUUGUAUGGCUUUGUUGGCCCAAAGCCAAAG ((((..((((.---------....(((.(((........))).)))(((((..(((((((.....)))))))..)))))))))..))))......((((((((......))))))))... ( -33.30) >consensus UAACCCGCACA_________AGCCGGUCA_ACUUUUCUCCAUCGGCAAAUGCACAAUUUGGCCUGCAAAAUGCAUAUUUGUGGCAUCCAUUUGUAUGGCUUUGUUGGUCCGU______AG .....................((((((.............)))))).............((((.(((((...(((((..((((...))))..)))))..))))).))))........... (-16.75 = -17.00 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:57 2006