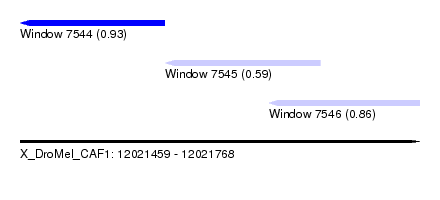
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,021,459 – 12,021,768 |
| Length | 309 |
| Max. P | 0.927719 |

| Location | 12,021,459 – 12,021,571 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -20.98 |
| Energy contribution | -22.34 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12021459 112 - 22224390 CUGGUUGAAAAGAUUGCUGCUUUUUGUAUACAAGAGGCAUGCAAAUGGUUUUUUAAUGGGGUUUCAGAUUUAUCAACAAAGUGAAACUGUAAACUAAAGUAAUAAAAAUUCC-------- ((.(((((((((.(((((((((((((....))))))))).))))....))))))))).))(((((((.(((((.......))))).))).))))..................-------- ( -28.00) >DroSec_CAF1 54783 120 - 1 CUGGUUGAAAAGAUUGCUGCUUUUUGUUUACAAGAGCCUUGCAAAUGGUUUUUUAAUGGGGUUUCAGAUUUAUCAACAAAGUGAAACUGUAAACUAAAGUAAUAAAAAUUCCGUGAAAUA ..((........((((((.......(((((((((((((........)))))))......(((((((..(((......))).)))))))))))))...)))))).......))........ ( -22.56) >DroSim_CAF1 53510 120 - 1 CUGGUUGAAAAGAUUGCUGCUUUUUGUUUACAAGAGCCAUGCAAAUGGUUUUUUAAUGGGGUUUCAGAUUUAUCAACAAAGUGAAACUGUAAACUAAAGUAAUAAAAAUUCCAUGAAAUA .(((........((((((.......(((((((((((((((....)))))))))......(((((((..(((......))).)))))))))))))...)))))).......)))....... ( -25.76) >DroEre_CAF1 51887 120 - 1 CUGUUUGAAAAGAUUGCUGCACUUUGUUUACAAACGGCAUGCAAAUGGUUUUUUAAUGGGGUUUCAGAUUUAUCAACAAAGUGAAACUGUAAACUAAAGUAAUAAAAAUUCCAUGGAACA ..((((.......(((((((..((((....))))..))).))))((((.((((((((..((((((((.(((((.......))))).))).)))))...))..))))))..)))).)))). ( -24.10) >DroYak_CAF1 53059 119 - 1 CUGGUUGAAAAGAUUGCAGCUUUUUGUUUACAAAAGGCAUGCAAAUGGUUUUUUAAUGGGGUUUCAGUUUUAAAAACAAAGUGAAACUGCAAACUAAAGUAAUAA-AAUUCCAUGGAAUA ((.(((((((((.(((((((((((((....)))))))).)))))....))))))))).))((((((((((((.........)))))))).))))...........-.((((....)))). ( -32.50) >consensus CUGGUUGAAAAGAUUGCUGCUUUUUGUUUACAAGAGGCAUGCAAAUGGUUUUUUAAUGGGGUUUCAGAUUUAUCAACAAAGUGAAACUGUAAACUAAAGUAAUAAAAAUUCCAUGAAAUA ((.(((((((((.(((((((((((((....))))))))).))))....))))))))).))(((((((.(((((.......))))).))).)))).......................... (-20.98 = -22.34 + 1.36)

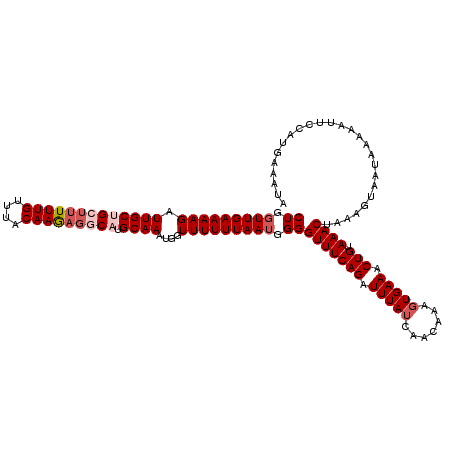
| Location | 12,021,571 – 12,021,691 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -17.55 |
| Energy contribution | -17.18 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12021571 120 - 22224390 AGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAUUUAACAACUAGCACAGUACAUUUUGUAUAGUAUCAAUUUGCAAAAGCAAAUAGGGAUUUUCGUUAAACAAAUGCAGAAUA ....((((((.(((((....))))).)))))).......((((((.((((..((((......)))).)))).((.((((((....)))))).)).......))))))............. ( -22.30) >DroSec_CAF1 54903 120 - 1 AGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAUUUAACAACUAGCACAGUACAUUUUGUAUAGUAUCAAUUUGCAAAAGCAAAUAGGAAUUUUCGUUAAACAAAUGCAAAAUA ....((((((.(((((....))))).)))))).......((((((.((((..((((......)))).)))).((.((((((....)))))).)).......))))))............. ( -22.30) >DroSim_CAF1 53630 120 - 1 AGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAUUUAACAACUAGCUCAGUACAUUUUGUAUAGUAUCAAUUUGCAAAAACAAAUAGGGAUUUUCGUUAAACAAAUGCAAAAUA ....((((((.(((((....))))).)))))).......((((((.((((...(((......)))..)))).((.(((((......))))).)).......))))))............. ( -17.40) >DroYak_CAF1 53178 120 - 1 AAAAUAGCUUCGUGGGGCAACUUGCUAAUUUAAAAUCAAUUUAACAACAAUGAUAGUAAAUUUUGUAUAGUAUCAAUUUGCAAAAGCAAAUAGGGAUUUUCGUUAAAUAAAUGCAAAAUA ......((...(..((....))..).............(((((((.....(((((.((.........)).)))))((((((....))))))..........)))))))....))...... ( -21.80) >consensus AGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAUUUAACAACUAGCACAGUACAUUUUGUAUAGUAUCAAUUUGCAAAAGCAAAUAGGGAUUUUCGUUAAACAAAUGCAAAAUA ....((((((.(((((....))))).)))))).......((((((.((((...(((......)))..)))).((.((((((....)))))).)).......))))))............. (-17.55 = -17.18 + -0.37)



| Location | 12,021,651 – 12,021,768 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12021651 117 - 22224390 AACAGUUUUUAUUGGCUAACUACCAAUUUUUACCACCUCUUUCAAUUUUUUUCAGAUUCUGAGAAAAAUUAAAAACGAGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAU ....(((((((((((.......)))))................((((((((((((...))))))))))))))))))((...((((((.(((((....))))).))))))...))... ( -24.80) >DroSec_CAF1 54983 116 - 1 AACAGUUUUUAUCGGCUAAUUACCAAUUUCUACCACCUCUUUCACU-UUUUUCAGAUACUGAGAAAAAUUAAAAACGAGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAU ....(((((((..((.......)).....................(-((((((((...)))))))))..)))))))((...((((((.(((((....))))).))))))...))... ( -21.10) >DroSim_CAF1 53710 116 - 1 AACCGUUUUUAUCGGCUAAUUACCAAUUUUUACCACCUCUUUCACU-UUUUUCAGAUUCUGAGAAAAAGUAAAAACGAGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAU ...(((((((((.((.......)).....................(-((((((((...))))))))).)))))))))....((((((.(((((....))))).))))))........ ( -22.50) >consensus AACAGUUUUUAUCGGCUAAUUACCAAUUUUUACCACCUCUUUCACU_UUUUUCAGAUUCUGAGAAAAAUUAAAAACGAGAAUGAAUUUGUAAGGGAGCUUACUAAUUUAAAAUCAAU ....(((((((..((.......)).......................((((((((...))))))))...)))))))((...((((((.(((((....))))).))))))...))... (-20.10 = -20.10 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:52 2006