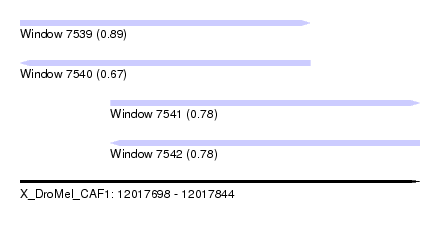
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,017,698 – 12,017,844 |
| Length | 146 |
| Max. P | 0.889339 |

| Location | 12,017,698 – 12,017,804 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12017698 106 + 22224390 AUUCAAGUGUGCUUGCUACAAUGCUGGCAUCGAAUGCAAGG---AGAUGGCGGGGUACAGCU-UACUGUACUAG--CGCAAGUACAGUGGAGCCAACAAUAGGCAAUGACAA .((((..(((((((((.......(((.((((..........---.)))).)))(((((((..-..)))))))..--.))))))))).))))(((.......)))........ ( -37.00) >DroSec_CAF1 51143 106 + 1 AUUCAAGUGUGCUCGCUACAAUGCUGGCAUCGAAUGCAAGG---AGAUGGCGGUGUACAGCU-UACUGUACUAG--UGCAAGUACAGUGGAGCCAACAAUGGGCAAUGACAA .((((..((((((.((.((...((((.((((..........---.)))).))))((((((..-..))))))..)--))).)))))).))))(((.......)))........ ( -33.60) >DroSim_CAF1 50336 109 + 1 AUUCAAGUGUGCUCGCUACAAUGCUGGCAUCGAAUGCAAGGAGGAGAUGGCGGGGCACAGCU-UACUGUACUAG--CGCAAGUACAGUGGAGCCAACAAUGGGCAAUGACAA .((((((((((((((((((..(.((.((((...)))).)).)...).)))).))))))).))-)((((((((..--....)))))))))))(((.......)))........ ( -33.60) >DroEre_CAF1 48245 106 + 1 AUUCAAGUGUGCUCGCUACAAUGCUGCCAUCGAAUGCACGG---AGAUGGCGGG-UGCAGCUUUACUGUACUAG--UGCCAGUACAGUGGAGCCGACAAUGGGCAAUGACAA ..(((....((((((........((((((((..........---.))))))))(-(.(.(((((((((((((..--....))))))))))))).)))..)))))).)))... ( -39.70) >DroYak_CAF1 49349 94 + 1 AUUCAAGUGUGCUGGCUACAAUGCUGCCAUCGAAUGCAAGG---AGAUGGCGGGUUGCAGCUUUACUGUACUAGCGUACAGGUACAGUGGAGCCGAC--------------- .((((..(((((.((((.((((.((((((((..........---.)))))))))))).))))...((((((....))))))))))).))))......--------------- ( -34.90) >consensus AUUCAAGUGUGCUCGCUACAAUGCUGGCAUCGAAUGCAAGG___AGAUGGCGGGGUACAGCU_UACUGUACUAG__UGCAAGUACAGUGGAGCCAACAAUGGGCAAUGACAA .......(((((((((......))((.((((..............)))).)))))))))(((.(((((((((........))))))))).)))................... (-25.24 = -25.36 + 0.12)

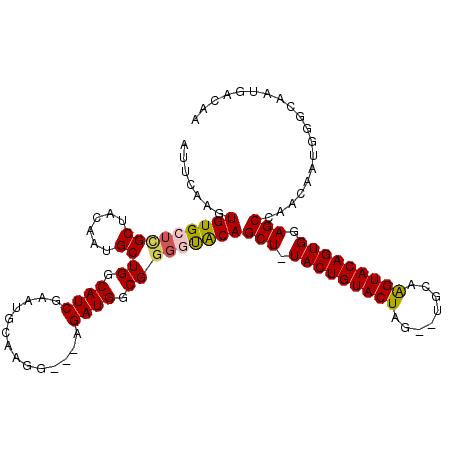

| Location | 12,017,698 – 12,017,804 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.54 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12017698 106 - 22224390 UUGUCAUUGCCUAUUGUUGGCUCCACUGUACUUGCG--CUAGUACAGUA-AGCUGUACCCCGCCAUCU---CCUUGCAUUCGAUGCCAGCAUUGUAGCAAGCACACUUGAAU .(((..((((.((((((((((((...((((..((.(--(..((((((..-..))))))...))))...---...))))...)).)))))))..)))))))..)))....... ( -27.40) >DroSec_CAF1 51143 106 - 1 UUGUCAUUGCCCAUUGUUGGCUCCACUGUACUUGCA--CUAGUACAGUA-AGCUGUACACCGCCAUCU---CCUUGCAUUCGAUGCCAGCAUUGUAGCGAGCACACUUGAAU .(((..((((.((.(((((((((.((((((((....--..)))))))).-.((.(....).)).....---..........)).))))))).))..))))..)))....... ( -28.10) >DroSim_CAF1 50336 109 - 1 UUGUCAUUGCCCAUUGUUGGCUCCACUGUACUUGCG--CUAGUACAGUA-AGCUGUGCCCCGCCAUCUCCUCCUUGCAUUCGAUGCCAGCAUUGUAGCGAGCACACUUGAAU .(((..((((.((.(((((((((.((((((((....--..)))))))).-....((((.................))))..)).))))))).))..))))..)))....... ( -28.23) >DroEre_CAF1 48245 106 - 1 UUGUCAUUGCCCAUUGUCGGCUCCACUGUACUGGCA--CUAGUACAGUAAAGCUGCA-CCCGCCAUCU---CCGUGCAUUCGAUGGCAGCAUUGUAGCGAGCACACUUGAAU .((.((........)).))((((.(((((((((...--.)))))))))...((((((-.(.((((((.---..........)))))).)...)))))))))).......... ( -33.80) >DroYak_CAF1 49349 94 - 1 ---------------GUCGGCUCCACUGUACCUGUACGCUAGUACAGUAAAGCUGCAACCCGCCAUCU---CCUUGCAUUCGAUGGCAGCAUUGUAGCCAGCACACUUGAAU ---------------....(((..(((((((..(....)..)))))))...(((((((..(((((((.---..........)))))).)..))))))).))).......... ( -28.50) >consensus UUGUCAUUGCCCAUUGUUGGCUCCACUGUACUUGCA__CUAGUACAGUA_AGCUGUACCCCGCCAUCU___CCUUGCAUUCGAUGCCAGCAUUGUAGCGAGCACACUUGAAU .................(((((..((((((((........))))))))..))))).................(((((...(((((....)))))..)))))........... (-22.30 = -22.54 + 0.24)


| Location | 12,017,731 – 12,017,844 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.04 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12017731 113 + 22224390 AUGCAAGG---AGAUGGCGGGGUACAGCU-UACUGUACUAG--CGCAAGUACAGUGGAGCCAACAAUAGGCAAUGACAAUGCGUUUAUAUGUUGAUGAUUUGCAGCCAUAGCAAAUUGU ..((((.(---..(((((...(((..(((-(((((((((..--....)))))))).))))(((((((((((.((....))..)))))).)))))......))).)))))..)...)))) ( -34.40) >DroSec_CAF1 51176 113 + 1 AUGCAAGG---AGAUGGCGGUGUACAGCU-UACUGUACUAG--UGCAAGUACAGUGGAGCCAACAAUGGGCAAUGACAAUGCGUUCAAAUGCUGAUGAUUUGCAGCCCAGACAAAUUGA ...(((..---.(.(((((((.....)))-(((((((((..--....)))))))))..)))).)..(((((..(((........)))..(((.........))))))))......))). ( -29.10) >DroSim_CAF1 50369 116 + 1 AUGCAAGGAGGAGAUGGCGGGGCACAGCU-UACUGUACUAG--CGCAAGUACAGUGGAGCCAACAAUGGGCAAUGACAAUGCGUUCAAAUGCUGAUGAUUUGCAGCCCAGACAAAUUGU ..............((...((((...(((-(((((((((..--....)))))))).)))).........((((.......((((....)))).......)))).))))...))...... ( -31.34) >DroEre_CAF1 48278 113 + 1 AUGCACGG---AGAUGGCGGG-UGCAGCUUUACUGUACUAG--UGCCAGUACAGUGGAGCCGACAAUGGGCAAUGACAAUGCCUGCAGAUGCUGGUGACUUGCAGCCAAGAGGAAUAAU .((((.(.---.(..((((..-(((.(((((((((((((..--....))))))))))))).......(((((.......))))))))..))))..)..).)))).((....))...... ( -39.50) >DroYak_CAF1 49382 94 + 1 AUGCAAGG---AGAUGGCGGGUUGCAGCUUUACUGUACUAGCGUACAGGUACAGUGGAGCCGAC----------------------AGAUGCUCAUGAUUUGCAGUUAAGGCAAAUUAU .((((((.---..((((((.((((..(((((((((((((........)))))))))))))))))----------------------...))).)))..))))))............... ( -32.90) >consensus AUGCAAGG___AGAUGGCGGGGUACAGCU_UACUGUACUAG__UGCAAGUACAGUGGAGCCAACAAUGGGCAAUGACAAUGCGUUCAAAUGCUGAUGAUUUGCAGCCAAGACAAAUUGU .((((((........((((.......(((.(((((((((........))))))))).))).........(((.......))).......)))).....))))))............... (-22.17 = -22.04 + -0.13)

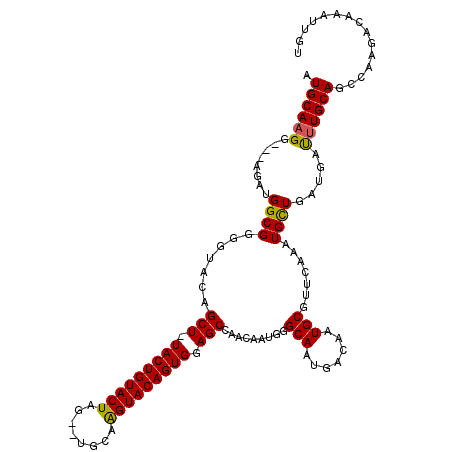

| Location | 12,017,731 – 12,017,844 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -18.03 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12017731 113 - 22224390 ACAAUUUGCUAUGGCUGCAAAUCAUCAACAUAUAAACGCAUUGUCAUUGCCUAUUGUUGGCUCCACUGUACUUGCG--CUAGUACAGUA-AGCUGUACCCCGCCAUCU---CCUUGCAU ......(((.(((((((((......(((((.(((...(((.......))).))))))))(((..((((((((....--..)))))))).-)))))))....)))))..---....))). ( -26.90) >DroSec_CAF1 51176 113 - 1 UCAAUUUGUCUGGGCUGCAAAUCAUCAGCAUUUGAACGCAUUGUCAUUGCCCAUUGUUGGCUCCACUGUACUUGCA--CUAGUACAGUA-AGCUGUACACCGCCAUCU---CCUUGCAU ......(((..((((((........))))........(((.......)))....((((((((..((((((((....--..)))))))).-))))).))).........---))..))). ( -25.40) >DroSim_CAF1 50369 116 - 1 ACAAUUUGUCUGGGCUGCAAAUCAUCAGCAUUUGAACGCAUUGUCAUUGCCCAUUGUUGGCUCCACUGUACUUGCG--CUAGUACAGUA-AGCUGUGCCCCGCCAUCUCCUCCUUGCAU ......((.(.((((.(((......(((((..((...(((.......))).)).)))))(((..((((((((....--..)))))))).-)))))))))).).)).............. ( -28.80) >DroEre_CAF1 48278 113 - 1 AUUAUUCCUCUUGGCUGCAAGUCACCAGCAUCUGCAGGCAUUGUCAUUGCCCAUUGUCGGCUCCACUGUACUGGCA--CUAGUACAGUAAAGCUGCA-CCCGCCAUCU---CCGUGCAU .............((((........))))...((((((((.......))))...((.(((((..(((((((((...--.)))))))))..)))))))-..........---...)))). ( -32.40) >DroYak_CAF1 49382 94 - 1 AUAAUUUGCCUUAACUGCAAAUCAUGAGCAUCU----------------------GUCGGCUCCACUGUACCUGUACGCUAGUACAGUAAAGCUGCAACCCGCCAUCU---CCUUGCAU ...............(((((...(((.((...(----------------------(.(((((..(((((((..(....)..)))))))..)))))))....)))))..---..))))). ( -24.10) >consensus ACAAUUUGUCUUGGCUGCAAAUCAUCAGCAUCUGAACGCAUUGUCAUUGCCCAUUGUUGGCUCCACUGUACUUGCA__CUAGUACAGUA_AGCUGUACCCCGCCAUCU___CCUUGCAU ...............(((((.................(((.......))).......(((((..((((((((........))))))))..)))))..................))))). (-18.03 = -18.34 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:48 2006