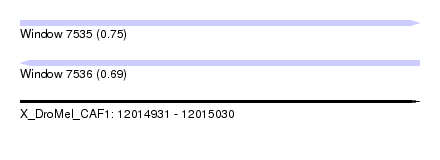
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,014,931 – 12,015,030 |
| Length | 99 |
| Max. P | 0.751448 |

| Location | 12,014,931 – 12,015,030 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -10.29 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
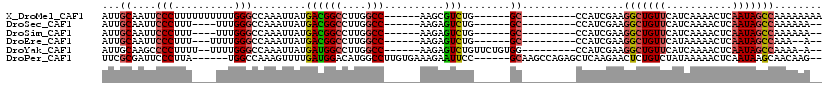
>X_DroMel_CAF1 12014931 99 + 22224390 AUUGCAAUUCCCUUUUUUUUUUGGGCCAAAUUAUGACGGCCUUGGCC------AAGCGUCUG------GC---------CCAUCGAAGGCUGUUCAUCAAAACUCAAUAGCCAAAAAAAA .............(((((((((((((((....(((..(((....)))------...))).))------))---------))).....(((((((...........))))))))))))))) ( -27.50) >DroSec_CAF1 48393 93 + 1 AUUGCAAUUCCCUUU----UUUGGGCCAAAUUAUGACGGCCUUGGCC------AAGAGUCUG------GC---------CCAUCGAAGGCUGUUCAUCAAAACUCAAUAGCCAAAAAA-- ............(((----..(((((((.(((.....(((....)))------...))).))------))---------)))..)))(((((((...........)))))))......-- ( -27.30) >DroSim_CAF1 47582 93 + 1 AUUGCAAUUCCCUUU----UUUGGGCCAAAUUAUGACGGCCUUGGCC------AAGAGUCUG------GC---------CCAUCGAAGGCUGUUCAUCAAAACUCAAUAGCCAAAAAA-- ............(((----..(((((((.(((.....(((....)))------...))).))------))---------)))..)))(((((((...........)))))))......-- ( -27.30) >DroEre_CAF1 45488 92 + 1 AUUGCAAUUCCCUUU---UUUUGGGCCAAAUUAUGACGGCCUUGGCC------AAGAGUCUG------GC---------CCAUCGAAGGCUGUUCAUAAAAACUCAAUAGCCAAA--A-- ...((....(((...---....))).....(((((((((((((((((------(......))------))---------).....))))))).))))))..........))....--.-- ( -27.40) >DroYak_CAF1 46590 100 + 1 AUUGCAAGCCCCUUUU--UUUUGGGCCAAAUUAUGAUGGCCUUGGCC------AAGAGUCUGUUCUGUGG---------CCAUCGAAGGCUGUUCAUCAAAACUCAAUAGCCAAAA-A-- .......((((.....--....)))).......((((((((..(..(------(......))..)...))---------))))))..(((((((...........)))))))....-.-- ( -28.30) >DroPer_CAF1 153237 106 + 1 UUCGCGAUUCCCUUA------UGGCCAAAGUUUUGAUGGACAUGGCCUUGUGAAAGAAUUCC------GCAAGCCAGAGCUCAAGAACUCUGUCUAUAAAAACUCAAUAAGCAACAAG-- ...((.....((...------.))....((((((.(((((((((((.(((((.........)------))))))))(((........)))))))))).))))))......))......-- ( -25.90) >consensus AUUGCAAUUCCCUUU____UUUGGGCCAAAUUAUGACGGCCUUGGCC______AAGAGUCUG______GC_________CCAUCGAAGGCUGUUCAUCAAAACUCAAUAGCCAAAAAA__ ...((....(((..........))).........((((((....)))..........)))........)).................(((((((...........)))))))........ (-10.29 = -10.52 + 0.22)

| Location | 12,014,931 – 12,015,030 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12014931 99 - 22224390 UUUUUUUUGGCUAUUGAGUUUUGAUGAACAGCCUUCGAUGG---------GC------CAGACGCUU------GGCCAAGGCCGUCAUAAUUUGGCCCAAAAAAAAAAGGGAAUUGCAAU ((((((((((...(..((((.(((((....(((((.....(---------((------(((....))------)))))))))))))).))))..).)))))))))).............. ( -29.40) >DroSec_CAF1 48393 93 - 1 --UUUUUUGGCUAUUGAGUUUUGAUGAACAGCCUUCGAUGG---------GC------CAGACUCUU------GGCCAAGGCCGUCAUAAUUUGGCCCAAA----AAAGGGAAUUGCAAU --.......((.(((..((((....))))..((((...(((---------((------((((....(------(((....))))......)))))))))..----.)))).))).))... ( -28.50) >DroSim_CAF1 47582 93 - 1 --UUUUUUGGCUAUUGAGUUUUGAUGAACAGCCUUCGAUGG---------GC------CAGACUCUU------GGCCAAGGCCGUCAUAAUUUGGCCCAAA----AAAGGGAAUUGCAAU --.......((.(((..((((....))))..((((...(((---------((------((((....(------(((....))))......)))))))))..----.)))).))).))... ( -28.50) >DroEre_CAF1 45488 92 - 1 --U--UUUGGCUAUUGAGUUUUUAUGAACAGCCUUCGAUGG---------GC------CAGACUCUU------GGCCAAGGCCGUCAUAAUUUGGCCCAAAA---AAAGGGAAUUGCAAU --.--....((.(((..((((....))))..((((...(((---------((------((((....(------(((....))))......)))))))))...---.)))).))).))... ( -27.80) >DroYak_CAF1 46590 100 - 1 --U-UUUUGGCUAUUGAGUUUUGAUGAACAGCCUUCGAUGG---------CCACAGAACAGACUCUU------GGCCAAGGCCAUCAUAAUUUGGCCCAAAA--AAAAGGGGCUUGCAAU --.-.....((....(((((.(((((....((((....(((---------(((.(((......))))------)))))))))))))).)))))(((((....--.....))))).))... ( -30.90) >DroPer_CAF1 153237 106 - 1 --CUUGUUGCUUAUUGAGUUUUUAUAGACAGAGUUCUUGAGCUCUGGCUUGC------GGAAUUCUUUCACAAGGCCAUGUCCAUCAAAACUUUGGCCA------UAAGGGAAUCGCGAA --(((((.(((....(((((((.((.(((((((((....)))))((((((..------.(((....)))...)))))))))).)).))))))).))).)------))))........... ( -31.70) >consensus __UUUUUUGGCUAUUGAGUUUUGAUGAACAGCCUUCGAUGG_________GC______CAGACUCUU______GGCCAAGGCCGUCAUAAUUUGGCCCAAA____AAAGGGAAUUGCAAU ......((((((.....((((....))))..((......))................................))))))(((((........)))))....................... (-13.63 = -13.55 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:43 2006