
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,014,413 – 12,014,542 |
| Length | 129 |
| Max. P | 0.999921 |

| Location | 12,014,413 – 12,014,504 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12014413 91 + 22224390 GACAAUUUGCCAAACAAUCUGUCAAGCCGGAAACAAUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACG ((((..(((.....)))..))))...(((....)..(((((((...(((((((.((....)))))((....)))))))))))))----------.)).... ( -24.10) >DroSec_CAF1 47859 91 + 1 GACAAUUUGCCAAACAAUCUGUCAAGCCGGAAACAAUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACG ((((..(((.....)))..))))...(((....)..(((((((...(((((((.((....)))))((....)))))))))))))----------.)).... ( -24.10) >DroSim_CAF1 47061 91 + 1 GACAAUUUGCCAAACAAUCUGUCAAGCCGGAAACAAUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACG ((((..(((.....)))..))))...(((....)..(((((((...(((((((.((....)))))((....)))))))))))))----------.)).... ( -24.10) >DroEre_CAF1 44950 91 + 1 GACAAUUUGCCAAACAAUCUGUCAAGCCGGAAACAAUUCGCACAACUUUCUCCAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACG ((((..(((.....)))..))))...(((....)..(((((((...((((....((....))...((....)))))))))))))----------.)).... ( -22.80) >DroYak_CAF1 46035 101 + 1 GACAAUUUGCCAAACAAUCUGUCAAGCCGGAAACAAUUCGCACAACUUUCUCUAGAGUAAUCAGAGUGCUCGCGAAAGUGCGAAUGGAACAGUACGGAACG ((((..(((.....)))..))))...(((....).((((((((...((((....(((((.......)))))..))))))))))))..........)).... ( -25.20) >consensus GACAAUUUGCCAAACAAUCUGUCAAGCCGGAAACAAUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA__________CGGAACG ((((..(((.....)))..))))...(((....)..(((((((.((((......)))).......((....))....)))))))...........)).... (-22.24 = -22.08 + -0.16)

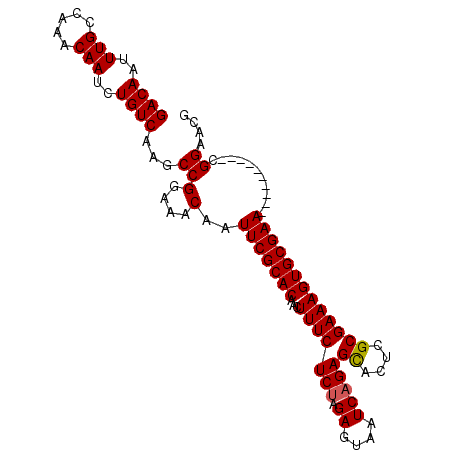
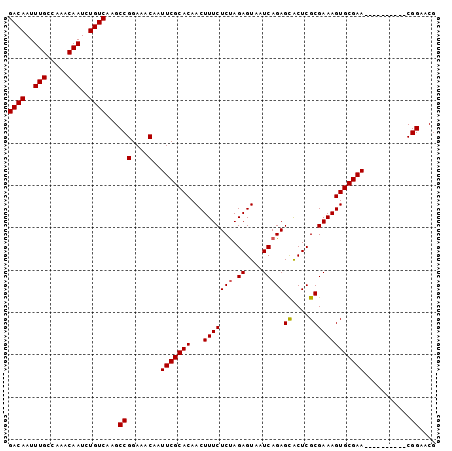
| Location | 12,014,448 – 12,014,542 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 93.93 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -27.92 |
| Energy contribution | -29.10 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12014448 94 + 22224390 AUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACGGAGAGUUUGCGAAAUAAUCAAGGGGGUACUUGAUGAGU-- .((((((.((((((((((.((....)))))(((((......)))))...----------.......)))))))))))))...((((((......))))))....-- ( -30.20) >DroSec_CAF1 47894 94 + 1 AUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACGGAGAGUUUGCGAAAUAAUCAAGGGGGUACUUGAUGAGU-- .((((((.((((((((((.((....)))))(((((......)))))...----------.......)))))))))))))...((((((......))))))....-- ( -30.20) >DroSim_CAF1 47096 94 + 1 AUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACGGAGAGUUUGCGAAAUAAUCAAGGGGGUACUUGAUGAGU-- .((((((.((((((((((.((....)))))(((((......)))))...----------.......)))))))))))))...((((((......))))))....-- ( -30.20) >DroEre_CAF1 44985 94 + 1 AUUCGCACAACUUUCUCCAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA----------CGGAACGGAGAGUUUGCGAAAUAAUCAAGGGGGUACUUGAUGAGU-- .((((((.((((((((((.((....))...(((((......)))))...----------.)))...)))))))))))))...((((((......))))))....-- ( -31.70) >DroYak_CAF1 46070 106 + 1 AUUCGCACAACUUUCUCUAGAGUAAUCAGAGUGCUCGCGAAAGUGCGAAUGGAACAGUACGGAACGGAGAGUUUGCGAAAUAAUCAAGGGGGUACUUGAUGAGUAG .((((((.((((((((((.((....)))))((((((((....)))..........)))))......)))))))))))))...((((((......))))))...... ( -31.50) >consensus AUUCGCACAACUUUCUCUAGAGUAAUCAGAGCACUCGCGAAAGUGCGAA__________CGGAACGGAGAGUUUGCGAAAUAAUCAAGGGGGUACUUGAUGAGU__ .((((((.((((((((...((....))...(((((......)))))...................))))))))))))))...((((((......))))))...... (-27.92 = -29.10 + 1.18)

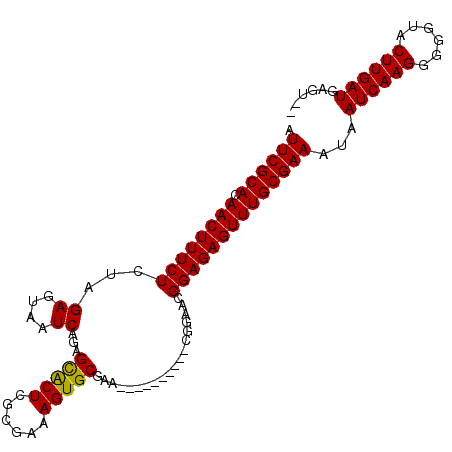
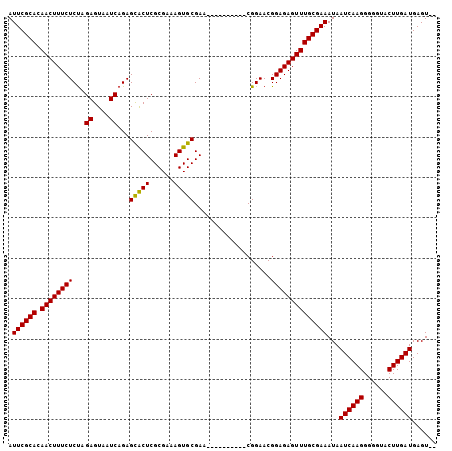
| Location | 12,014,448 – 12,014,542 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 93.93 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12014448 94 - 22224390 --ACUCAUCAAGUACCCCCUUGAUUAUUUCGCAAACUCUCCGUUCCG----------UUCGCACUUUCGCGAGUGCUCUGAUUACUCUAGAGAAAGUUGUGCGAAU --....((((((......))))))...((((((((((.((.......----------...((((((....))))))((((.......)))))).)))).)))))). ( -22.60) >DroSec_CAF1 47894 94 - 1 --ACUCAUCAAGUACCCCCUUGAUUAUUUCGCAAACUCUCCGUUCCG----------UUCGCACUUUCGCGAGUGCUCUGAUUACUCUAGAGAAAGUUGUGCGAAU --....((((((......))))))...((((((((((.((.......----------...((((((....))))))((((.......)))))).)))).)))))). ( -22.60) >DroSim_CAF1 47096 94 - 1 --ACUCAUCAAGUACCCCCUUGAUUAUUUCGCAAACUCUCCGUUCCG----------UUCGCACUUUCGCGAGUGCUCUGAUUACUCUAGAGAAAGUUGUGCGAAU --....((((((......))))))...((((((((((.((.......----------...((((((....))))))((((.......)))))).)))).)))))). ( -22.60) >DroEre_CAF1 44985 94 - 1 --ACUCAUCAAGUACCCCCUUGAUUAUUUCGCAAACUCUCCGUUCCG----------UUCGCACUUUCGCGAGUGCUCUGAUUACUCUGGAGAAAGUUGUGCGAAU --....((((((......))))))...(((((((((((((((.....----------.((((((((....))))))...))......))))))..))).)))))). ( -25.50) >DroYak_CAF1 46070 106 - 1 CUACUCAUCAAGUACCCCCUUGAUUAUUUCGCAAACUCUCCGUUCCGUACUGUUCCAUUCGCACUUUCGCGAGCACUCUGAUUACUCUAGAGAAAGUUGUGCGAAU ......((((((......))))))...((((((((((....((((((...(((.......)))....)).)))).(((((.......)))))..)))).)))))). ( -22.50) >consensus __ACUCAUCAAGUACCCCCUUGAUUAUUUCGCAAACUCUCCGUUCCG__________UUCGCACUUUCGCGAGUGCUCUGAUUACUCUAGAGAAAGUUGUGCGAAU ......((((((......))))))...((((((((((....................(((((......)))))..(((((.......)))))..)))).)))))). (-21.70 = -21.54 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:41 2006