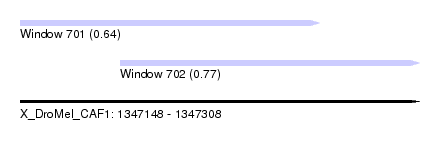
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,347,148 – 1,347,308 |
| Length | 160 |
| Max. P | 0.772782 |

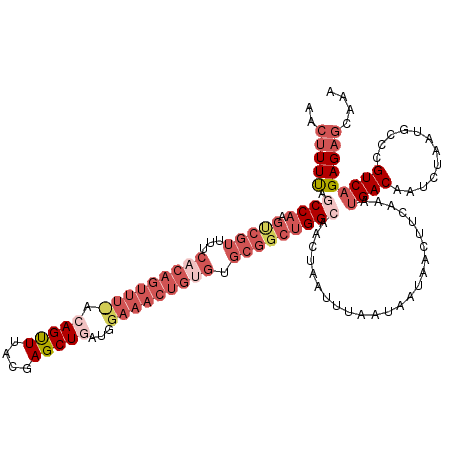
| Location | 1,347,148 – 1,347,268 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -19.88 |
| Energy contribution | -23.70 |
| Covariance contribution | 3.82 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1347148 120 + 22224390 GCAACAGUGAAAAGUUUCCCACCCAUACUUGACGCGCUUUAACUUUUAGCCAAGUCGUUUUCACAGUUUUACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUA ......(((..((((...........))))..))).............((((.(((((...(((((((((.(((((....)))))...))))))))).)))))))))............. ( -33.70) >DroSec_CAF1 7567 120 + 1 GCAACAGUGAAAAGUUUGCCACCCAUACUUGACGCGCUUUAACUUUUAGCCAAGUCGUUUUCACAGUUUCACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUA .....(((.(((((((.((....((....))....))...))))))).((((.(((((...(((((((((.(((((....)))))...))))))))).))))))))).)))......... ( -37.30) >DroSim_CAF1 10790 120 + 1 GCAACAGUGAAAAGUUUGCCACCCAUACUUGACGCGCUUUAACUUUUAGCCAAGUCGUUUUCACAGUUUCACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUA .....(((.(((((((.((....((....))....))...))))))).((((.(((((...(((((((((.(((((....)))))...))))))))).))))))))).)))......... ( -37.30) >DroEre_CAF1 7622 95 + 1 GCAACGGCGAAAAGUUUGCCACCCACACUUGAUGCGCUUCAACUUUCAGCCAAGCCGUUUUCAC--------AGUUUACGAGCUGAU-----------------GGAAACUAAUUUAAUA .....(((..((((((.((((..((....)).)).))...))))))..))).....((((((((--------((((....))))).)-----------------)))))).......... ( -22.60) >DroYak_CAF1 8035 112 + 1 GCCACAGUGAAAAGUUUGCCACCCACACUUGAUGCGCUUUAACUUUUAGCCAAGCCGUUUUCCC--------AGCUUACGAGCUGAUGCAAGCUGUGUGCGGCUGGAAACUAAUUUAAUA ............(((((.((((((((((....((((((.........))).............(--------((((....)))))..)))....))))).)).))))))))......... ( -27.70) >consensus GCAACAGUGAAAAGUUUGCCACCCAUACUUGACGCGCUUUAACUUUUAGCCAAGUCGUUUUCACAGUUU_ACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUA .....((((..((((.((.....)).))))....))))..........((((.(((((...(((((((((.(((((....)))))...))))))))).)))))))))............. (-19.88 = -23.70 + 3.82)

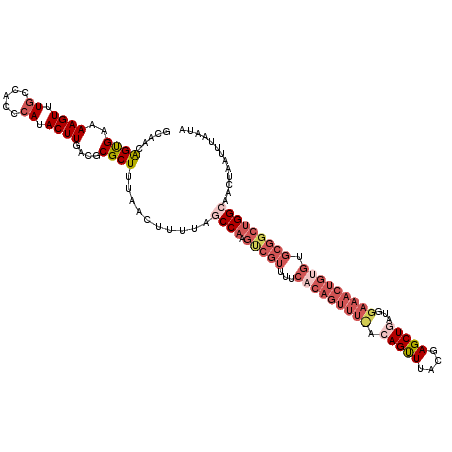
| Location | 1,347,188 – 1,347,308 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.98 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -17.46 |
| Energy contribution | -21.84 |
| Covariance contribution | 4.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1347188 120 + 22224390 AACUUUUAGCCAAGUCGUUUUCACAGUUUUACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUAAUAACUUCAAAUGACAAUCUAAUGCCCGUCAGAGAGCAAA ..(((((.((((.(((((...(((((((((.(((((....)))))...))))))))).)))))))))........................((((............))))))))).... ( -33.80) >DroSec_CAF1 7607 114 + 1 AACUUUUAGCCAAGUCGUUUUCACAGUUUCACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUAAUAACUUCAAAUGACAAUCUAAUGCCCGUCAGAG------ ........((((.(((((...(((((((((.(((((....)))))...))))))))).)))))))))........................((((............))))...------ ( -33.50) >DroSim_CAF1 10830 120 + 1 AACUUUUAGCCAAGUCGUUUUCACAGUUUCACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUAAUAACUUCAAAUGACAAUCUAAUGCCCGUCAGAGAGCAAA ..(((((.((((.(((((...(((((((((.(((((....)))))...))))))))).)))))))))........................((((............))))))))).... ( -35.60) >DroEre_CAF1 7662 95 + 1 AACUUUCAGCCAAGCCGUUUUCAC--------AGUUUACGAGCUGAU-----------------GGAAACUAAUUUAAUAAUAAAUUCAAAUGACAAUCUAAUGCCCGUCAGAGAGUGGA .((((((.........((((((((--------((((....))))).)-----------------)))))).....................((((............))))))))))... ( -19.50) >DroYak_CAF1 8075 112 + 1 AACUUUUAGCCAAGCCGUUUUCCC--------AGCUUACGAGCUGAUGCAAGCUGUGUGCGGCUGGAAACUAAUUUAAUAAUAACUUCAAACGACAAUCUAAUGCCCGUCAGAGAGUGAA .((((((..(((.(((((...(.(--------(((((....((....)))))))).).))))))))..........................(((............))).))))))... ( -25.70) >consensus AACUUUUAGCCAAGUCGUUUUCACAGUUU_ACAGUUUACGAGCUGAUGGAAACUGUGUGCGGCUGGCAACUAAUUUAAUAAUAACUUCAAAUGACAAUCUAAUGCCCGUCAGAGAGCAAA ..(((((.((((.(((((...(((((((((.(((((....)))))...))))))))).)))))))))........................((((............))))))))).... (-17.46 = -21.84 + 4.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:01 2006