
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,009,901 – 12,010,101 |
| Length | 200 |
| Max. P | 0.986919 |

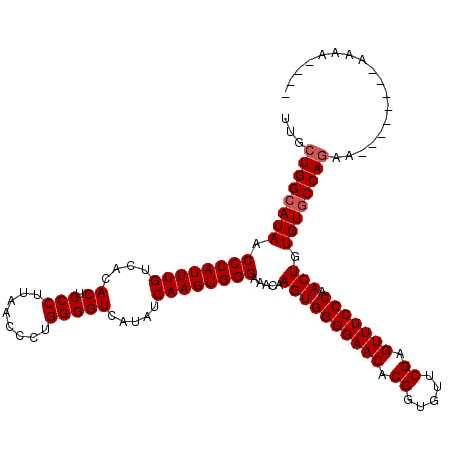
| Location | 12,009,901 – 12,010,021 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -39.17 |
| Energy contribution | -39.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.32 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12009901 120 + 22224390 UUGCUGGCAUAACCCAUUUGUCACACUCCCUUAACCCUGGGGUCAUAUCAAGUGGGAAACAAGUGGGGAACACGGUAUUCGAGUUUCCCAACUGUGUACCAAAACAAGAAAAAAAAACAA ....(((.....((((((((....((.(((........))))).....))))))))..(((..((((((((.((.....)).))))))))..)))...)))................... ( -34.80) >DroSec_CAF1 43341 108 + 1 UUGCUGGCAUAACCCAUUUGUCACACUCCCUUAACCCUGGGGUCAUAUCAAGUGGGGAACAAGUGGGGAACACGGCGUUCGAGUUUCCCCACUGUGUGCCAGAA--------AAAA---- ...((((((((.((((((((....((.(((........))))).....)))))))).....((((((((((.((.....)).).))))))))).))))))))..--------....---- ( -46.00) >DroSim_CAF1 42601 108 + 1 UUGCUGGCAUAACCCAUUUGUCACACUCCCUUAACCCUGGGGUCAUAUCAAGUGGGAAACAAGUGGGGAACACGGUGUUCGAGUUUCCCAACUGUGUGCCAGAA--------AAAA---- ...(((((((..((((((((....((.(((........))))).....))))))))..(((..((((((((.((.....)).))))))))..))))))))))..--------....---- ( -44.20) >consensus UUGCUGGCAUAACCCAUUUGUCACACUCCCUUAACCCUGGGGUCAUAUCAAGUGGGAAACAAGUGGGGAACACGGUGUUCGAGUUUCCCAACUGUGUGCCAGAA________AAAA____ ...((((((((.((((((((....((.(((........))))).....)))))))).....((((((((((.((.....)).))))))).))).)))))))).................. (-39.17 = -39.83 + 0.67)

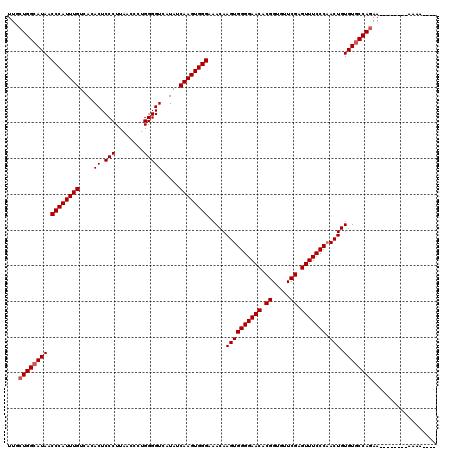
| Location | 12,009,901 – 12,010,021 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -33.41 |
| Energy contribution | -33.30 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12009901 120 - 22224390 UUGUUUUUUUUUCUUGUUUUGGUACACAGUUGGGAAACUCGAAUACCGUGUUCCCCACUUGUUUCCCACUUGAUAUGACCCCAGGGUUAAGGGAGUGUGACAAAUGGGUUAUGCCAGCAA ..............(((..(((((.((((.((((.(((.((.....)).))).)))).))))..((((.(((.((((..(((........)))..)))).))).))))...)))))))). ( -31.90) >DroSec_CAF1 43341 108 - 1 ----UUUU--------UUCUGGCACACAGUGGGGAAACUCGAACGCCGUGUUCCCCACUUGUUCCCCACUUGAUAUGACCCCAGGGUUAAGGGAGUGUGACAAAUGGGUUAUGCCAGCAA ----....--------..((((((((.(((((((((...((.....))..))))))))))))..((((.(((.((((..(((........)))..)))).))).))))....)))))... ( -40.30) >DroSim_CAF1 42601 108 - 1 ----UUUU--------UUCUGGCACACAGUUGGGAAACUCGAACACCGUGUUCCCCACUUGUUUCCCACUUGAUAUGACCCCAGGGUUAAGGGAGUGUGACAAAUGGGUUAUGCCAGCAA ----....--------..((((((.((((.((((.(((.((.....)).))).)))).))))..((((.(((.((((..(((........)))..)))).))).))))...))))))... ( -35.20) >consensus ____UUUU________UUCUGGCACACAGUUGGGAAACUCGAACACCGUGUUCCCCACUUGUUUCCCACUUGAUAUGACCCCAGGGUUAAGGGAGUGUGACAAAUGGGUUAUGCCAGCAA ..................((((((.((((.((((.(((.((.....)).))).)))).))))..((((.(((.((((..(((........)))..)))).))).))))...))))))... (-33.41 = -33.30 + -0.11)

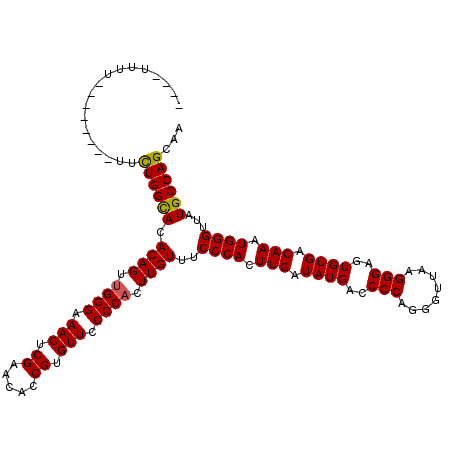

| Location | 12,009,941 – 12,010,061 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -24.11 |
| Energy contribution | -23.45 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12009941 120 + 22224390 GGUCAUAUCAAGUGGGAAACAAGUGGGGAACACGGUAUUCGAGUUUCCCAACUGUGUACCAAAACAAGAAAAAAAAACAAAACAAAAAAAACUUUAAAAUACGCUUUCCACUCCACAUGC ...(((....(((((((((((..((((((((.((.....)).))))))))..)))((......)).......................................))))))))....))). ( -23.80) >DroSec_CAF1 43381 104 + 1 GGUCAUAUCAAGUGGGGAACAAGUGGGGAACACGGCGUUCGAGUUUCCCCACUGUGUGCCAGAA--------AAAA--------AUAAAAACUUUAAUACACGCUUUCCACUCCACAUGC ...........((((((....((((((((((.((.....)).).)))))))))(((((....((--------(...--------........)))....)))))......)))))).... ( -29.40) >DroSim_CAF1 42641 102 + 1 GGUCAUAUCAAGUGGGAAACAAGUGGGGAACACGGUGUUCGAGUUUCCCAACUGUGUGCCAGAA--------AAAA--------ACA--AACUUUGAAAUACGCUUUCCACUUCACAUGC ...(((.(.(((((((((.(.((((((((((.((.....)).))))))).)))((((..((((.--------....--------...--...))))..))))).))))))))).).))). ( -27.50) >consensus GGUCAUAUCAAGUGGGAAACAAGUGGGGAACACGGUGUUCGAGUUUCCCAACUGUGUGCCAGAA________AAAA________AAAAAAACUUUAAAAUACGCUUUCCACUCCACAUGC ...(((....((((((((...((((((((((.((.....)).))))))).)))(((((.........................................)))))))))))))....))). (-24.11 = -23.45 + -0.66)

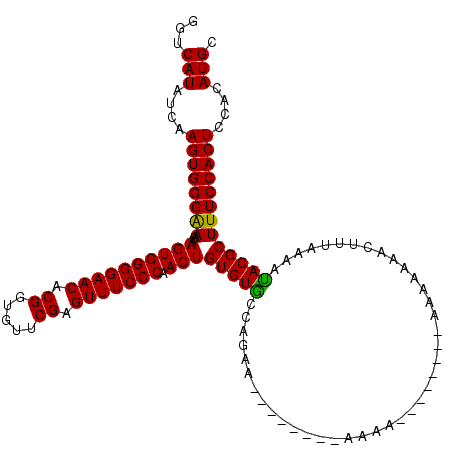
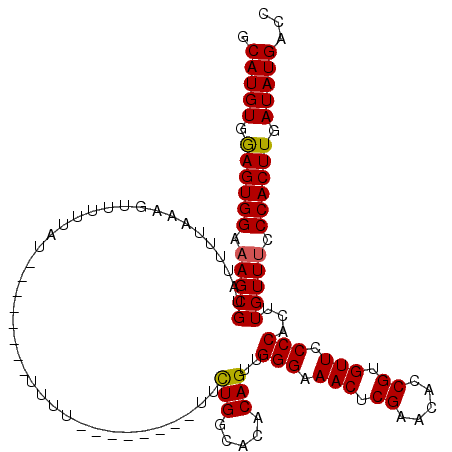
| Location | 12,009,941 – 12,010,061 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.83 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12009941 120 - 22224390 GCAUGUGGAGUGGAAAGCGUAUUUUAAAGUUUUUUUUGUUUUGUUUUUUUUUCUUGUUUUGGUACACAGUUGGGAAACUCGAAUACCGUGUUCCCCACUUGUUUCCCACUUGAUAUGACC .(((((.((((((.(((((((((...(((.......................))).....)))))..(((.(((((...((.....))..))))).))).)))).)))))).)))))... ( -30.00) >DroSec_CAF1 43381 104 - 1 GCAUGUGGAGUGGAAAGCGUGUAUUAAAGUUUUUAU--------UUUU--------UUCUGGCACACAGUGGGGAAACUCGAACGCCGUGUUCCCCACUUGUUCCCCACUUGAUAUGACC .(((((.((((((..(((((((...((((.......--------.)))--------)....))))..(((((((((...((.....))..))))))))).)))..)))))).)))))... ( -38.10) >DroSim_CAF1 42641 102 - 1 GCAUGUGAAGUGGAAAGCGUAUUUCAAAGUU--UGU--------UUUU--------UUCUGGCACACAGUUGGGAAACUCGAACACCGUGUUCCCCACUUGUUUCCCACUUGAUAUGACC .(((((.((((((.(((((.........(((--((.--------..((--------(((..((.....))..)))))..)))))...(((.....))).))))).)))))).)))))... ( -31.00) >consensus GCAUGUGGAGUGGAAAGCGUAUUUUAAAGUUUUUAU________UUUU________UUCUGGCACACAGUUGGGAAACUCGAACACCGUGUUCCCCACUUGUUUCCCACUUGAUAUGACC .(((((.((((((.(((((.......................................(((.....)))..(((.(((.((.....)).))).)))...))))).)))))).)))))... (-27.94 = -27.83 + -0.11)


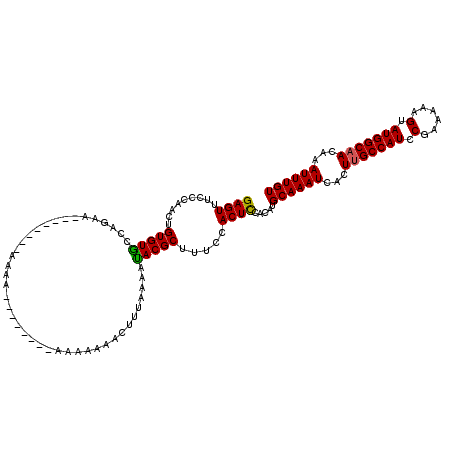
| Location | 12,009,981 – 12,010,101 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -14.76 |
| Consensus MFE | -13.18 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12009981 120 + 22224390 GAGUUUCCCAACUGUGUACCAAAACAAGAAAAAAAAACAAAACAAAAAAAACUUUAAAAUACGCUUUCCACUCCACAUGCAAAUCACUCGCCAUUCCAAAAAGUAUGGCAACAAAUUUGU ((((........(((........)))....................................((..............)).....))))(((((.(......).)))))........... ( -11.14) >DroSec_CAF1 43421 104 + 1 GAGUUUCCCCACUGUGUGCCAGAA--------AAAA--------AUAAAAACUUUAAUACACGCUUUCCACUCCACAUGCAAAUCACUUGCCAUCCGAAAAAGUAUGGCAACAAAUUUGU ((((.........(((((....((--------(...--------........)))....))))).....)))).....((((((...(((((((.(......).)))))))...)))))) ( -17.04) >DroSim_CAF1 42681 102 + 1 GAGUUUCCCAACUGUGUGCCAGAA--------AAAA--------ACA--AACUUUGAAAUACGCUUUCCACUUCACAUGCAAAUCACUUGCCAUCCGAAAAAGUAUGGCAACAAAUUUGU .((((....))))((((..((((.--------....--------...--...))))..))))................((((((...(((((((.(......).)))))))...)))))) ( -16.10) >consensus GAGUUUCCCAACUGUGUGCCAGAA________AAAA________AAAAAAACUUUAAAAUACGCUUUCCACUCCACAUGCAAAUCACUUGCCAUCCGAAAAAGUAUGGCAACAAAUUUGU ((((.........(((((.........................................))))).....)))).....((((((...(((((((.(......).)))))))...)))))) (-13.18 = -12.85 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:35 2006