
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,999,145 – 11,999,398 |
| Length | 253 |
| Max. P | 0.909466 |

| Location | 11,999,145 – 11,999,249 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.45 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11999145 104 + 22224390 CUUCUACGCAUAUAAAUUUGUAUGCGUUGCACUGAAAUUUGCAACAGUUACAAUUACAACGAUAAAAGCCGGUGGCAGCAACAGCAAA---CAUGCAACCAUGCCCA .......(((((((....)))))))((((((........))))))..............((........))(.((((......(((..---..))).....))))). ( -21.90) >DroSec_CAF1 38125 87 + 1 ---GUACGCAUAUAAAUUUGUAUGUGUGGCACUGAAAAUUGCAACAGUUACAAUUACAACGAUAAAAGCCGG-GGC------AGCAAC---CAUGCAA-------CA ---.((((((((((....))))))))))(((.((....((((....(((........))).......((...-.))------.)))).---)))))..-------.. ( -17.80) >DroEre_CAF1 36864 83 + 1 -----ACGCAUAUAAAUUUGUAUGCGUAGCAGUCGAAUUUGCAACAGUUACAAUUACAACGAUAAAGGCCG--GGC------GGCAAC---CAUGCCAC-------- -----(((((((((....))))))))).((((......))))........................(((..--((.------.....)---)..)))..-------- ( -19.80) >DroYak_CAF1 37596 90 + 1 -----GCGCAUAUAAAAUUGUAUGCGUCGCAGUGAAAUUUGCAACAGUUACAAUUACAACGAUAAAAGCUGG-GGC------AGCAACACACAUGCCACCA-----A -----(((((((((....))))))))).((((((....((((..(((((...(((.....)))...))))).-.))------)).....))).))).....-----. ( -23.80) >consensus _____ACGCAUAUAAAUUUGUAUGCGUAGCACUGAAAUUUGCAACAGUUACAAUUACAACGAUAAAAGCCGG_GGC______AGCAAC___CAUGCAAC_______A .....(((((((((....))))))))).(((.......((((....(((........))).......(((...))).......))))......)))........... (-17.01 = -16.45 + -0.56)

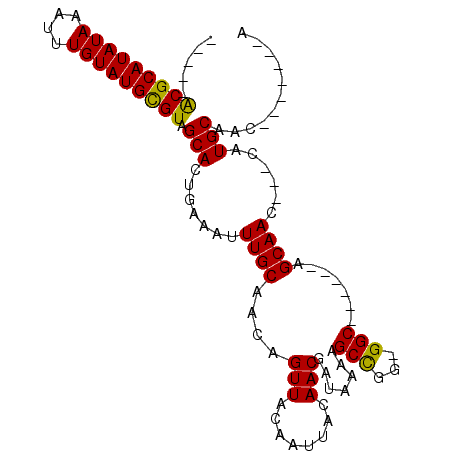

| Location | 11,999,249 – 11,999,365 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -17.61 |
| Energy contribution | -18.11 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11999249 116 - 22224390 CUUUUUUUUGGGCCAUGUCGUGUGUUCAUUUGUCUGUGUGCUCGUUUCAUCGUUUGGCUUUUGCCAUUUGACUGAGCGCAUAAU----AAAAAUUGGCACAUAUUUUUGGUUUCAGUGUG .......((((((((....((((((..((((...((((((((((.....(((..((((....))))..))).))))))))))..----..))))..)))))).....))).))))).... ( -35.40) >DroSec_CAF1 38212 114 - 1 CUCUU-UUUGGGCCACGUCGUGUGUUCAUUUGUCUGUGUGCA-GCGGCAUCGUUUGGCUUUUGCCAUUUGACUGAGCGCACAAU----AAAAAUUGGCACAUAUUUUUGGUUUCAGUGUG .....-.((((((((....((((((..((((...(((((((.-.(((..(((..((((....))))..)))))).)))))))..----..))))..)))))).....))).))))).... ( -34.20) >DroEre_CAF1 36947 87 - 1 -------------------------UCAUUUGCCUG--UGCA-GCGGCAUCGUUCGGCUUUUGCCGUUUGACUGAGCGCAUAAU----AAAAAUUGGCACAUAUUUUUGGUUUCAGUGU- -------------------------.((((.(((((--(((.-(((.(((((..((((....))))..))).))..))).((((----....))))))))).......)))...)))).- ( -23.11) >DroYak_CAF1 37686 116 - 1 CUUU--UUUGGGCCAUGUCGUAUGUUCAUUUGUGUGAAUGCA-GCGACAUCGUUUGGCUUUUGCCGUUUGACUGAGCGUAUAAUAUGUAAAAAUUGGCACAUAUUUUUGGUUU-GGUGUG ....--...((((.(((((((..((((((....))))))...-))))))).))))(((....))).....((..(((....(((((((..........)))))))....))).-.))... ( -29.60) >consensus CUUU__UUUGGGCCAUGUCGUGUGUUCAUUUGUCUGUGUGCA_GCGGCAUCGUUUGGCUUUUGCCAUUUGACUGAGCGCAUAAU____AAAAAUUGGCACAUAUUUUUGGUUUCAGUGUG ...................(((((((((...((......))........(((..((((....))))..))).)))))))))...............((((...............)))). (-17.61 = -18.11 + 0.50)


| Location | 11,999,285 – 11,999,398 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -27.40 |
| Energy contribution | -26.63 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11999285 113 - 22224390 -------CAUUUUAACCGCGACUGUCCAAACGGUUAGCCACUUUUUUUUGGGCCAUGUCGUGUGUUCAUUUGUCUGUGUGCUCGUUUCAUCGUUUGGCUUUUGCCAUUUGACUGAGCGCA -------((..(.(((((((((.(((((((.(((.....)))....)))))))...)))))).))).)..))....((((((((.....(((..((((....))))..))).)))))))) ( -35.20) >DroSec_CAF1 38248 111 - 1 -------CAUUUUAACCGCGGCUGUCUAAACGGUUAGCCACUCUU-UUUGGGCCACGUCGUGUGUUCAUUUGUCUGUGUGCA-GCGGCAUCGUUUGGCUUUUGCCAUUUGACUGAGCGCA -------..........(((((.(((((((.((.........)).-)))))))...)))))(((((((...(((.(((.(((-(.(((........))).)))))))..)))))))))). ( -31.90) >DroYak_CAF1 37725 117 - 1 ACCUGUGCGUUUAAACCGCCGCUGUCCAAAGGGUUAGCUACUUU--UUUGGGCCAUGUCGUAUGUUCAUUUGUGUGAAUGCA-GCGACAUCGUUUGGCUUUUGCCGUUUGACUGAGCGUA .....((((((((....(((((((.((....)).))))..(...--...))))..((((((..((((((....))))))...-))))))(((..((((....))))..))).)))))))) ( -35.40) >consensus _______CAUUUUAACCGCGGCUGUCCAAACGGUUAGCCACUUUU_UUUGGGCCAUGUCGUGUGUUCAUUUGUCUGUGUGCA_GCGACAUCGUUUGGCUUUUGCCAUUUGACUGAGCGCA .................(((((.(((((((.(((.....)))....)))))))...)))))(((((((...((......))........(((..((((....))))..))).))))))). (-27.40 = -26.63 + -0.77)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:29 2006