
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,994,828 – 11,994,940 |
| Length | 112 |
| Max. P | 0.995419 |

| Location | 11,994,828 – 11,994,940 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -43.81 |
| Consensus MFE | -31.01 |
| Energy contribution | -33.33 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11994828 112 + 22224390 GUGUGUGUUUAGGAGGAGGAGCGGAUGCAGGGGCAUGUGAUGGCACUUGCCCUUUGACCCCUGCCCCUUUUGCCCCUCAUUGUUUGGGGCACAACAAAAAGCAAAUAAAGAU .(((.(((((...((((((.((((..(((((((((.(((....))).))))).))).)..)))).))))))(((((.........)))))........))))).)))..... ( -39.30) >DroSec_CAF1 33845 112 + 1 GUGUGUGUUUAGGAGGGGGAGGGGCUUCAGGGGCAUGUGAUGGCACUUGCCCUUCGACCCCUGCCCCUUUGGCCCCUCAUUGUUUGGGGUACAACAAAAAGCAAAUAAAGAC .(((.(((((.(((((((.(((((.((.(((((((.(((....))).))))))).))))))).))))))).(((((.........)))))........))))).)))..... ( -50.80) >DroEre_CAF1 32714 107 + 1 G----GGGAUGGGGG-GGGGGAGGGUGCAAGGGCACGUGAUGGCACUUGCCCUUUGACCCCUGCCCCAUUGGCCCCUCAUUGUUUGGAGUACAACAAAAAGCAAAUAAAGAU (----(((.....((-((.((.((((..(((((((.(((....))).)))))))..)))))).)))).....))))((.(((((((...............))))))).)). ( -46.56) >DroYak_CAF1 33875 111 + 1 GUGUGUGUUUAGGAG-AGGGGAGGGUGCAAGGGCAUGUGAUGGCACUUGCCCUUUGACCCCUGCCCCAUUUGCCCCUCAUUGUUUGGGAUACAACAAAAAGCAAAUAAAGAU .(((((.....(.((-(((((((((..((((((((.(((....))).))))).)))..)))).)))).))).)(((.........))))))))................... ( -38.60) >consensus GUGUGUGUUUAGGAG_AGGAGAGGGUGCAAGGGCAUGUGAUGGCACUUGCCCUUUGACCCCUGCCCCAUUGGCCCCUCAUUGUUUGGGGUACAACAAAAAGCAAAUAAAGAU ......((((.(((((((.((.((((..(((((((.(((....))).)))))))..)))))).))))))).(((((.........)))))........)))).......... (-31.01 = -33.33 + 2.31)

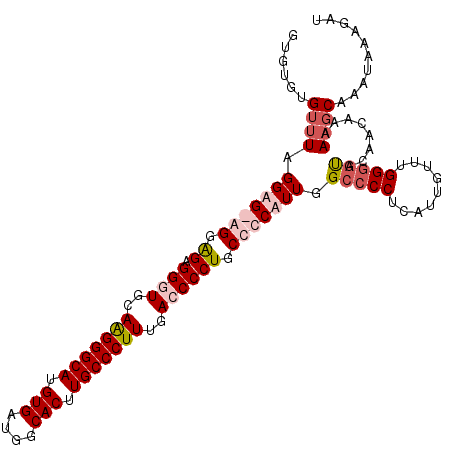
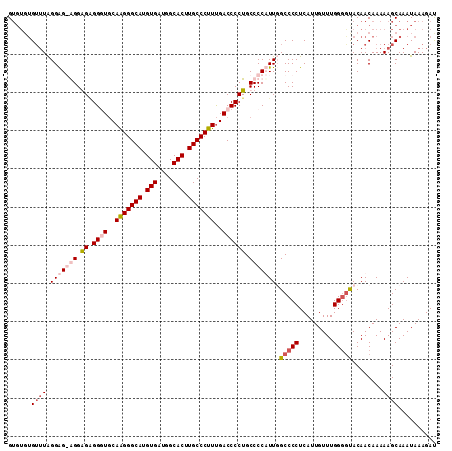
| Location | 11,994,828 – 11,994,940 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -36.34 |
| Energy contribution | -37.40 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11994828 112 - 22224390 AUCUUUAUUUGCUUUUUGUUGUGCCCCAAACAAUGAGGGGCAAAAGGGGCAGGGGUCAAAGGGCAAGUGCCAUCACAUGCCCCUGCAUCCGCUCCUCCUCCUAAACACACAC .......(((((((((..((((.......))))..)))))))))(((((.((((((....(((((.(((....))).)))))........)))))).))))).......... ( -39.50) >DroSec_CAF1 33845 112 - 1 GUCUUUAUUUGCUUUUUGUUGUACCCCAAACAAUGAGGGGCCAAAGGGGCAGGGGUCGAAGGGCAAGUGCCAUCACAUGCCCCUGAAGCCCCUCCCCCUCCUAAACACACAC ((.((((...((((((..((((.......))))..))))))...(((((.((((((....(((((.(((....))).))))).....)))))).)))))..)))).)).... ( -43.00) >DroEre_CAF1 32714 107 - 1 AUCUUUAUUUGCUUUUUGUUGUACUCCAAACAAUGAGGGGCCAAUGGGGCAGGGGUCAAAGGGCAAGUGCCAUCACGUGCCCUUGCACCCUCCCCC-CCCCCAUCCC----C ..........((((((..((((.......))))..))))))..((((((..((((...(((((((.(((....))).))))))).......)))).-.))))))...----. ( -41.90) >DroYak_CAF1 33875 111 - 1 AUCUUUAUUUGCUUUUUGUUGUAUCCCAAACAAUGAGGGGCAAAUGGGGCAGGGGUCAAAGGGCAAGUGCCAUCACAUGCCCUUGCACCCUCCCCU-CUCCUAAACACACAC ......((((((((((..((((.......))))..))))))))))((((.((((....(((((((.(((....))).)))))))...)))))))).-............... ( -45.20) >consensus AUCUUUAUUUGCUUUUUGUUGUACCCCAAACAAUGAGGGGCAAAAGGGGCAGGGGUCAAAGGGCAAGUGCCAUCACAUGCCCCUGCACCCUCCCCC_CUCCUAAACACACAC .......(((((((((..((((.......))))..))))))))).((((.(((((...(((((((.(((....))).))))))).......))))).))))........... (-36.34 = -37.40 + 1.06)

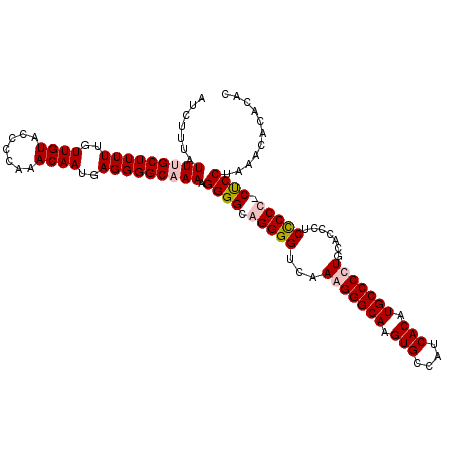
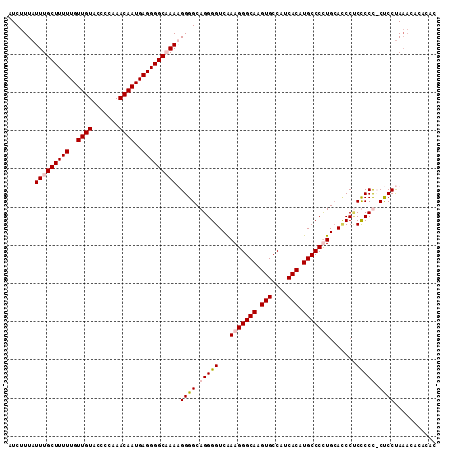
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:24 2006