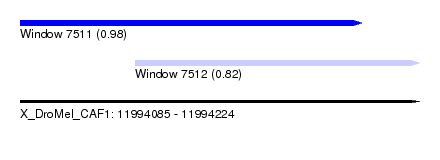
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,994,085 – 11,994,224 |
| Length | 139 |
| Max. P | 0.981973 |

| Location | 11,994,085 – 11,994,204 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -24.76 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
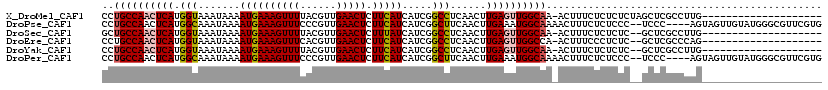
>X_DroMel_CAF1 11994085 119 + 22224390 UUUAUUAAUCAUUGGGGCUCCAGUUUUUGCCAGCAAUUUUCCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUUACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCAA-ACUU ..........((((.(((..........)))..)))).....((((((((((.(((.......((((((((((......))))).))))).....)))......)))))))))).-.... ( -29.70) >DroPse_CAF1 130943 119 + 1 UUUAUUAAUCAUUCGGGCUCCAGUU-UUGCCAGCAAUUUUCCUGCCAACUCAUGGCAAAUAAAAUGAAAGUUUCCCGUUGAACUCUUCAUCAUCGGCUUCAACUUGAAAUGGCAAAACUU .....................((((-(((((..((.(((...(((((.....)))))...))).))...(((((..((((((..(.........)..))))))..)))))))))))))). ( -27.30) >DroSec_CAF1 33177 119 + 1 UUUAUUAAUCAUUGGGGCUCCAGUUUUUGCCAGCAAUUUUGCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUUACGUUGAACUCUUUAUCAUCGGCCUCAACUUGAGUUGGCAA-ACUU ...............(((..........)))((((....))))(((((((((.(((...((((((....))))))((.(((........))).)))))......)))))))))..-.... ( -30.00) >DroEre_CAF1 32004 119 + 1 UUUAUUAAUCAUUGGGGCUCCAGUUUUUGCCUGCAAUUUUCCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUCACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCCA-ACUU ..........((((((((..........)))).))))......(((((((((.(((.......((((((((((......))))).))))).....)))......)))))))))..-.... ( -27.80) >DroYak_CAF1 33192 119 + 1 UUUAUUAAUCAUUGGGGCUCCAGUUUUUGCCGGCAAUUUUCCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUUACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCAA-ACUU ..........(((((....))))).((((((((((.......))))((((((.(((.......((((((((((......))))).))))).....)))......)))))))))))-)... ( -29.70) >DroPer_CAF1 135751 119 + 1 UUUAUUAAUCAUUCGGGCUCCAGUU-UUGCCAGCAAUUUUCCUGCCAACUCAUGGCAAAUAAAAUGAAAGUUUCCCGUUGAACUCUUCAUCAUCGGCUUCAACUUGAAAUGGCAAAACUU .....................((((-(((((..((.(((...(((((.....)))))...))).))...(((((..((((((..(.........)..))))))..)))))))))))))). ( -27.30) >consensus UUUAUUAAUCAUUGGGGCUCCAGUUUUUGCCAGCAAUUUUCCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUCACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCAA_ACUU ...............(((..........)))...........((((((((((.(((.......((((((((((......))))).))))).....)))......))))))))))...... (-24.76 = -25.02 + 0.25)


| Location | 11,994,125 – 11,994,224 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
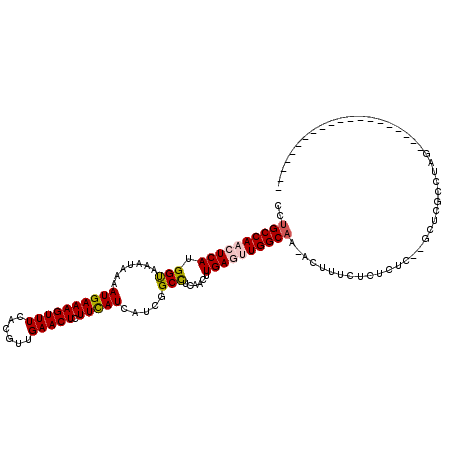
>X_DroMel_CAF1 11994125 99 + 22224390 CCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUUACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCAA-ACUUUCUCUCUCUAGCUCGCCUUG-------------------- ..((((((((((.(((.......((((((((((......))))).))))).....)))......)))))))))).-........................-------------------- ( -24.30) >DroPse_CAF1 130982 114 + 1 CCUGCCAACUCAUGGCAAAUAAAAUGAAAGUUUCCCGUUGAACUCUUCAUCAUCGGCUUCAACUUGAAAUGGCAAAACUUUCUCUCCC--UCCC----AGUAGUUGUAUGGGCGUUCGUG ..(((((.....)))))........((((((((.(((.(((........))).))).(((.....)))......)))))))).....(--.(((----(.........)))).)...... ( -21.60) >DroSec_CAF1 33217 97 + 1 GCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUUACGUUGAACUCUUUAUCAUCGGCCUCAACUUGAGUUGGCAA-ACUUUCUCUCUC--GCUCGCCUUG-------------------- ..((((((((((.(((...((((((....))))))((.(((........))).)))))......)))))))))).-............--..........-------------------- ( -24.10) >DroEre_CAF1 32044 97 + 1 CCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUCACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCCA-ACUUUCCCUCUC--GCUCGCCCAG-------------------- ...(((((((((.(((.......((((((((((......))))).))))).....)))......)))))))))..-............--..........-------------------- ( -21.80) >DroYak_CAF1 33232 97 + 1 CCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUUACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCAA-ACUUUCUCUCUC--GCUCGCCUUG-------------------- ..((((((((((.(((.......((((((((((......))))).))))).....)))......)))))))))).-............--..........-------------------- ( -24.30) >DroPer_CAF1 135790 114 + 1 CCUGCCAACUCAUGGCAAAUAAAAUGAAAGUUUCCCGUUGAACUCUUCAUCAUCGGCUUCAACUUGAAAUGGCAAAACUUUCUCUCCC--UCCC----AGUAGUUGUAUGGGCGUUCGUG ..(((((.....)))))........((((((((.(((.(((........))).))).(((.....)))......)))))))).....(--.(((----(.........)))).)...... ( -21.60) >consensus CCUGCCAACUCAUGGUAAAUAAAAUGAAAGUUUCACGUUGAACUCUUCAUCAUCGGCCUCAACUUGAGUUGGCAA_ACUUUCUCUCUC__GCUCGCCUAG____________________ ..((((((((((.(((.......((((((((((......))))).))))).....)))......)))))))))).............................................. (-20.95 = -21.20 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:22 2006