
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,989,701 – 11,989,878 |
| Length | 177 |
| Max. P | 0.997853 |

| Location | 11,989,701 – 11,989,810 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11989701 109 - 22224390 GCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGUCCAAAUGCAAAUGGGCUGUCAUAAUUAGCCCGCCCACACGAGUAC---CGCUUAUCAUUU--C------GACCCCCCAGC ......(((((.(((((((((......)))))))))..(((.(((((.....((((((.......))))))........(((...---..)))..)))))--.------)))..))))). ( -27.00) >DroPse_CAF1 126694 105 - 1 GCAUAAUUAGGAUUAAUUAUGCAUAAUCGUGAUUAACAGUCAACAUGCAAAUGGCCUGUCAUAAUUAGCCAUCCC-------------CGCUU--CAUCCGCCAUCCUCCAUCCGCCAUC ((......(((((......(((((.....((((.....))))..))))).(((((..((....))..)))))...-------------.....--........)))))......)).... ( -15.90) >DroSim_CAF1 30245 98 - 1 GCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGUCCAAAUGCAAAUGGGCUGUCAUAAUUAGCCCGCCCACACGAGUAC---CGCUUAUCAUUU--C----------------- ((((..(((((.(((((((((......)))))))))...)))))))))....((((((.......))))))........(((...---..))).......--.----------------- ( -24.20) >DroEre_CAF1 27977 100 - 1 GCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGUCCAAAUGCAAAUGUGCUGUCAUAAUUAGCCCGCCCACUCGAGUACCGCCGCUUAUCAUUC-------------------- ((......(((.((((((((((((....)))....((((..((........))..))))))))))))))))((..((....))...)).)).........-------------------- ( -18.50) >DroYak_CAF1 29153 103 - 1 GCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGUCCAAAUGCAAAUGUGCUGUCAUAAUUAGCCCGCCCACACGAGCAC---CGCUUAUCAUUC--C------AUUCU------ ((......(((.((((((((((((....)))....((((..((........))..))))))))))))))))((.(....).))..---.)).........--.------.....------ ( -19.70) >DroPer_CAF1 131520 91 - 1 GCAUAAUUAGGAUUAAUUAUGCAUAAUUGUGAUUAACAGUCAACAUGCAAAUGGCCUGUCAUAAUUAGCCAUCCC-------------CGCUU--CAUCCGCC--------------AUC (((((((((....))))))))).((((((((((.....((((.........))))..))))))))))........-------------.....--........--------------... ( -17.40) >consensus GCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGUCCAAAUGCAAAUGGGCUGUCAUAAUUAGCCCGCCCACACGAGUAC___CGCUUAUCAUUC__C_________________ (((((((((....))))))))).......((((((((((((((........))))))))..))))))..................................................... (-15.15 = -15.93 + 0.78)

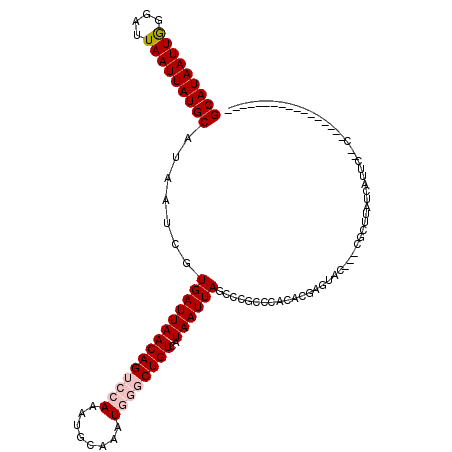

| Location | 11,989,770 – 11,989,878 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -17.57 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11989770 108 + 22224390 ACUGUUAAUCACGAUUAUGCAUAAUUAAUCCCAAUUAUGCACAAACACAAAUUAAUAGACAAAAUAUGCACAUUGCAGCUGUCGCAUCAAUUAAGCCGGUAGCCUG-------UA .((((((((........(((((((((......))))))))).........))))))))(((.....(((.....)))(((..(((.........).))..))).))-------). ( -21.93) >DroSec_CAF1 29085 108 + 1 ACUGUUAAUCACGAUUAUGCAUAAUUAAUCCCAAUUAUGCACAAACACAAAUUAAUAGACAAAAUAUGCACAUUGCAGCUGUCACAUCAAUUAAGCCGCUAGCCUG-------UA .((((((((........(((((((((......))))))))).........))))))))........((((....((.(((.............))).)).....))-------)) ( -18.75) >DroSim_CAF1 30303 108 + 1 ACUGUUAAUCACGAUUAUGCAUAAUUAAUCCCAAUUAUGCACAAACACAAAUUAAUAGACAAAAUAUGCACAUUGCAGCUGUCGCAUCAAUUAAGCCGCUCGCCUG-------UA .((((((((........(((((((((......))))))))).........))))))))........((((....((.(((.............))).)).....))-------)) ( -18.75) >DroEre_CAF1 28037 115 + 1 ACUGUUAAUCACGAUUAUGCAUAAUUAAUCCCAAUUAUGCACAAACACAAAUUAAUAGACAAAAUAUGCACAUUGCAGCUGUCGCAUCAAUUAAGCCGGUAGCCUAUUGCCCAUA .((((((((........(((((((((......))))))))).........))))))))........(((.....)))(((..(((.........).))..)))............ ( -21.23) >DroYak_CAF1 29216 108 + 1 ACUGUUAAUCACGAUUAUGCAUAAUUAAUCCCAAUUAUGCACAAACACAAAUUAAUAGACAAAAUAUGCACAUUGCAGCUGUCGCAUCAAUUAAGCCGGUAGCCUG-------UA .((((((((........(((((((((......))))))))).........))))))))(((.....(((.....)))(((..(((.........).))..))).))-------). ( -21.93) >consensus ACUGUUAAUCACGAUUAUGCAUAAUUAAUCCCAAUUAUGCACAAACACAAAUUAAUAGACAAAAUAUGCACAUUGCAGCUGUCGCAUCAAUUAAGCCGGUAGCCUG_______UA .((((((((........(((((((((......))))))))).........))))))))........(((.....)))((((((((.........).)))))))............ (-17.57 = -18.37 + 0.80)


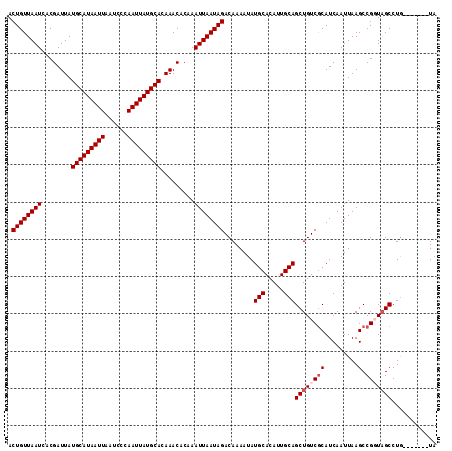
| Location | 11,989,770 – 11,989,878 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11989770 108 - 22224390 UA-------CAGGCUACCGGCUUAAUUGAUGCGACAGCUGCAAUGUGCAUAUUUUGUCUAUUAAUUUGUGUUUGUGCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGU ..-------.((((.....)))).(((((((.(((((.(((.....)))....))))))))))))(..((.(((((((((((((....))))))))))))).))..)........ ( -26.50) >DroSec_CAF1 29085 108 - 1 UA-------CAGGCUAGCGGCUUAAUUGAUGUGACAGCUGCAAUGUGCAUAUUUUGUCUAUUAAUUUGUGUUUGUGCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGU ..-------.((((.....)))).(((((((.(((((.(((.....)))....))))))))))))(..((.(((((((((((((....))))))))))))).))..)........ ( -26.50) >DroSim_CAF1 30303 108 - 1 UA-------CAGGCGAGCGGCUUAAUUGAUGCGACAGCUGCAAUGUGCAUAUUUUGUCUAUUAAUUUGUGUUUGUGCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGU ..-------...((((((((((.............))))))..((((((((.((.((((......((((((....))))))...)))).)).)))))))).)))).......... ( -28.02) >DroEre_CAF1 28037 115 - 1 UAUGGGCAAUAGGCUACCGGCUUAAUUGAUGCGACAGCUGCAAUGUGCAUAUUUUGUCUAUUAAUUUGUGUUUGUGCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGU .(((((((((((((.....)))))....((((.(((.......)))))))...))))))))....(..((.(((((((((((((....))))))))))))).))..)........ ( -30.70) >DroYak_CAF1 29216 108 - 1 UA-------CAGGCUACCGGCUUAAUUGAUGCGACAGCUGCAAUGUGCAUAUUUUGUCUAUUAAUUUGUGUUUGUGCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGU ..-------.((((.....)))).(((((((.(((((.(((.....)))....))))))))))))(..((.(((((((((((((....))))))))))))).))..)........ ( -26.50) >consensus UA_______CAGGCUACCGGCUUAAUUGAUGCGACAGCUGCAAUGUGCAUAUUUUGUCUAUUAAUUUGUGUUUGUGCAUAAUUGGGAUUAAUUAUGCAUAAUCGUGAUUAACAGU ..........((((.....)))).(((((((.(((((.(((.....)))....))))))))))))(..((.(((((((((((((....))))))))))))).))..)........ (-26.44 = -26.44 + -0.00)


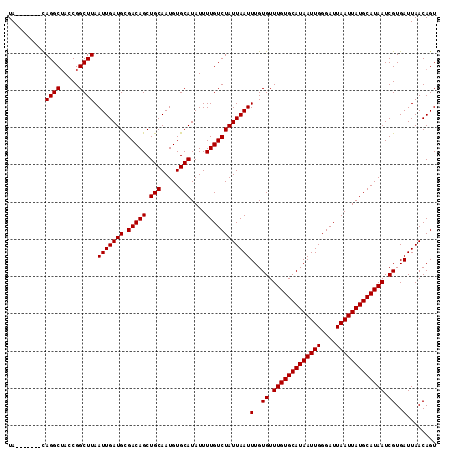
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:15 2006