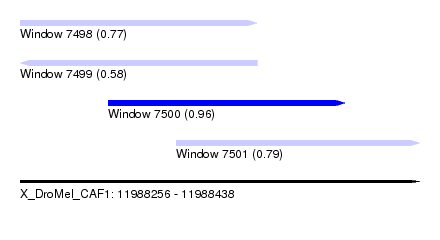
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,988,256 – 11,988,438 |
| Length | 182 |
| Max. P | 0.960804 |

| Location | 11,988,256 – 11,988,364 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.38 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11988256 108 + 22224390 AUUUCCAUCACUGCGGCAACAUCUCAACCAACAUUUUCAACGCCG---CC--ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAACGGGGGCGUUGCAACCGCACUAU ...........(((((..................((.(((((((.---((--((...........)).((.(((.......))).))....)).))))))).))))))).... ( -20.16) >DroSec_CAF1 27616 110 + 1 AUUUCCAUCACUGUGGAAACAUCUCAACCAACAUUUUCAACGCCACCACC--ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGU .(((((((....)))))))..................(((((((......--........((((((..((.(((.......))).)))))))-))))))))............ ( -21.03) >DroSim_CAF1 28849 110 + 1 AUUUCCAUCACUGCGGCAACAUCUCAACCAACAUUUUCAACGCCACCACC--ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGU ........((.(((((..................((.(((((((......--........((((((..((.(((.......))).)))))))-)))))))).))))))).)). ( -20.89) >DroEre_CAF1 26541 112 + 1 AUUUCCAUCACUGCGGCAACAUCUCAACCAACAUUUUCAGCGCCACCACUAACAUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGU ........((.(((((..................((.(((((((................((((((..((.(((.......))).)))))))-)))))))).))))))).)). ( -20.75) >DroYak_CAF1 27682 106 + 1 AUUUCCAUCAUUGCGGUAACAUCUCAACCAACAUUUUCAACGCCACU-----CGUAAAUUCCUUUGUAGCCAAAUCAAAUUUUCCGCCA-AG-GGGCGUUGCAACCGCACAGU ...........((((((....................(((((((...-----..........((((....)))).........((....-.)-))))))))..)))))).... ( -18.75) >consensus AUUUCCAUCACUGCGGCAACAUCUCAACCAACAUUUUCAACGCCACCACC__ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG_GGGCGUUGCAACCGCACUGU ...........(((((..................((.(((((((...........(((((..((((....))))...)))))............))))))).))))))).... (-15.70 = -15.38 + -0.32)

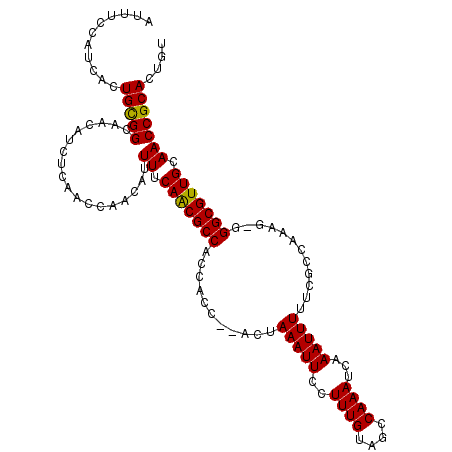
| Location | 11,988,256 – 11,988,364 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -23.96 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11988256 108 - 22224390 AUAGUGCGGUUGCAACGCCCCCGUUGGCGAAAAAUUUGAUUUGGCUACAAAGGAAUUUAGU--GG---CGGCGUUGAAAAUGUUGGUUGAGAUGUUGCCGCAGUGAUGGAAAU ......(.((..(..((((......))))..((((((..((((....)))).)))))).((--((---(((((((...(((....)))..))))))))))).)..)).).... ( -31.10) >DroSec_CAF1 27616 110 - 1 ACAGUGCGGUUGCAACGCCC-CUUUGGCGAAAAAUUUGAUUUGGCUACAAAGGAAUUUAGU--GGUGGUGGCGUUGAAAAUGUUGGUUGAGAUGUUUCCACAGUGAUGGAAAU .(((.(((....((((((((-(..(.((...((((((..((((....)))).)))))).))--.).)).)))))))....)))...)))....(((((((......))))))) ( -28.60) >DroSim_CAF1 28849 110 - 1 ACAGUGCGGUUGCAACGCCC-CUUUGGCGAAAAAUUUGAUUUGGCUACAAAGGAAUUUAGU--GGUGGUGGCGUUGAAAAUGUUGGUUGAGAUGUUGCCGCAGUGAUGGAAAU .((.((((((..((((((((-(..(.((...((((((..((((....)))).)))))).))--.).)).)))))))..((((((......)))))))))))).))........ ( -34.10) >DroEre_CAF1 26541 112 - 1 ACAGUGCGGUUGCAACGCCC-CUUUGGCGAAAAAUUUGAUUUGGCUACAAAGGAAUUUAUGUUAGUGGUGGCGCUGAAAAUGUUGGUUGAGAUGUUGCCGCAGUGAUGGAAAU .((.((((((.(((.(((((-(.((((((..((((((..((((....)))).)))))).)))))).)).)))((..(.....)..))...).))).)))))).))........ ( -33.50) >DroYak_CAF1 27682 106 - 1 ACUGUGCGGUUGCAACGCCC-CU-UGGCGGAAAAUUUGAUUUGGCUACAAAGGAAUUUACG-----AGUGGCGUUGAAAAUGUUGGUUGAGAUGUUACCGCAAUGAUGGAAAU ....((((((..(((((((.-((-((.....((((((..((((....)))).)))))).))-----)).)))))))..((((((......))))))))))))........... ( -29.50) >consensus ACAGUGCGGUUGCAACGCCC_CUUUGGCGAAAAAUUUGAUUUGGCUACAAAGGAAUUUAGU__GGUGGUGGCGUUGAAAAUGUUGGUUGAGAUGUUGCCGCAGUGAUGGAAAU .((.((((((.(((.((((......))))..((((((..((((....)))).))))))..........((((((.....)))))).......))).)))))).))........ (-23.96 = -24.32 + 0.36)



| Location | 11,988,296 – 11,988,404 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11988296 108 + 22224390 CGCCG---CC--ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAACGGGGGCGUUGCAACCGCACUAUGGCCACGCCCACUUUUGGCCAGCACACCGAAAUCCAUUAA .((.(---(.--((...........)).))...............(((((.((((((((.((............)).)))))).)).)))))..))................. ( -25.10) >DroSec_CAF1 27656 110 + 1 CGCCACCACC--ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGUGGCCACGCCCACUAUUGGCCAGCACACCGAAAUCCAUUAA .((((.....--.........(((((..((.(((.......))).)))))))-((((((.((.((......)).)).)))))).....))))..................... ( -24.70) >DroSim_CAF1 28889 110 + 1 CGCCACCACC--ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGUGGCCACGCCCACUUUUGGCCAGCACACCGAAAUCCAUUAA .((((.....--.........(((((..((.(((.......))).)))))))-((((((.((.((......)).)).)))))).....))))..................... ( -24.70) >DroEre_CAF1 26581 111 + 1 CGCCACCACUAACAUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGUGGCCACGCCCCC-CUUGGCCAGCACACCGAAAUCCAUUAA .((...................((((....))))...........(((((.(-((((((.((.((......)).)).))))))).-.)))))..))................. ( -26.30) >DroYak_CAF1 27722 106 + 1 CGCCACU-----CGUAAAUUCCUUUGUAGCCAAAUCAAAUUUUCCGCCA-AG-GGGCGUUGCAACCGCACAGUGGCAACGCCCACUUUUGGCCAGCACACCAAAAUCCAUUAA .((....-----..........((((....))))...........((((-(.-(((((((((.((......)).)))))))))....)))))..))................. ( -26.90) >consensus CGCCACCACC__ACUAAAUUCCUUUGUAGCCAAAUCAAAUUUUUCGCCAAAG_GGGCGUUGCAACCGCACUGUGGCCACGCCCACUUUUGGCCAGCACACCGAAAUCCAUUAA .((...................((((....))))...........(((((...((((((.((.((......)).)).))))))....)))))..))................. (-21.36 = -21.76 + 0.40)


| Location | 11,988,327 – 11,988,438 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -15.10 |
| Energy contribution | -17.49 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11988327 111 + 22224390 AA---AUUUUUCGCCAACGGGGGCGUUGCAACCGCACUAUGGCCACGCCCACUUUUGGCCAGCACACCGAAAUCCAUUAAGUGCAUUUUGCAAAAGAACUAACACUC------GAAUUCC ..---....((((((((.((((((((.((............)).)))))).)).)))))..((((...............)))).......................------))).... ( -27.46) >DroSec_CAF1 27690 109 + 1 AA---AUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGUGGCCACGCCCACUAUUGGCCAGCACACCGAAAUCCAUUAAGUGCAUUUUG-AAAAGAACUCAGACUC------GCAUUCC ..---.(((((((((((((-((((((.((.((......)).)).)))))).)).)))))..((((...............)))).....)-)))))...........------....... ( -30.46) >DroSim_CAF1 28923 109 + 1 AA---AUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGUGGCCACGCCCACUUUUGGCCAGCACACCGAAAUCCAUUAAGUGCAUUUUG-AAAAGAACUCACACUC------GCAUUCC ..---.(((((((((((((-((((((.((.((......)).)).)))))).).))))))..((((...............)))).....)-)))))...........------....... ( -30.46) >DroEre_CAF1 26617 114 + 1 AA---AUUUUUCGCCAAAG-GGGCGUUGCAACCGCACUGUGGCCACGCCCCC-CUUGGCCAGCACACCGAAAUCCAUUAAGUGCAUUGUG-AAAAGAACUCACACUCCUUCCAUGUUUCC ..---.......(((((.(-((((((.((.((......)).)).))))))).-.)))))..((((...............))))..((((-(.......)))))................ ( -32.76) >DroYak_CAF1 27753 108 + 1 AA---AUUUUCCGCCA-AG-GGGCGUUGCAACCGCACAGUGGCAACGCCCACUUUUGGCCAGCACACCAAAAUCCAUUAAGUGCAUUUUG-AAAAGAACUCACACUC------UCUUUCC ..---.(((((.((((-(.-(((((((((.((......)).)))))))))....)))))..((((...............)))).....)-))))............------....... ( -31.66) >DroPer_CAF1 129225 90 + 1 AACCAGUUUUGUGGCUGCGGGAG---GGGAAUCGCACUGCGGCCACGCCCACU--UGGC---------------CAUUAAGUGCAUUUCA-AA---CUCUCAGUCUC------UGAUUCC ...(((......(((((..((((---.(((((.(((((..(((((........--))))---------------)....)))))))))).-..---))))))))).)------))..... ( -30.30) >consensus AA___AUUUUUCGCCAAAG_GGGCGUUGCAACCGCACUGUGGCCACGCCCACUUUUGGCCAGCACACCGAAAUCCAUUAAGUGCAUUUUG_AAAAGAACUCACACUC______GCAUUCC ............(((((...((((((.((.((......)).)).))))))....)))))..((((...............)))).................................... (-15.10 = -17.49 + 2.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:11 2006