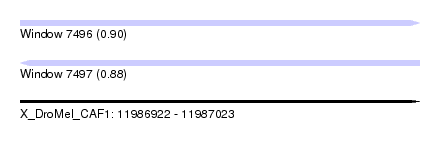
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,986,922 – 11,987,023 |
| Length | 101 |
| Max. P | 0.898166 |

| Location | 11,986,922 – 11,987,023 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11986922 101 + 22224390 CAGCCCCAUCCUUUUUUU-UUU-----------UUUUUUGGCCAAAACUUGCCAGCCAUUUUGGCCAGGACAACACCCUCGUCUUGGAAUUUUAUUUGCCACCUUUCUGGGCC ..((((............-...-----------.....((((........))))(((.....)))((((((.........))))))......................)))). ( -21.30) >DroSec_CAF1 26308 106 + 1 GAGCCCCAUCCAUAUUUU-UUUC------GCCCUUUUUUGGCCAAAACUUGCCAGCCAUUUUGGCCAGGACAACACCCUCGUCUUGGAAUUUUAUUUGCCACCUUUCUGGGCC ..................-....------((((.....((((........(((((.....)))))((((((.........))))))...........)))).......)))). ( -23.30) >DroSim_CAF1 26914 107 + 1 CAGCCCCAUCCAUAUUUUUUUUC------GCCCUUUUUUGGCCAAAACUUGCCAGCCAUUUUGGCCAGGACAACACCCUCGUCUUGGAAUUUUAUUUGCCACCUUUCUGGGCC .......................------((((.....((((........(((((.....)))))((((((.........))))))...........)))).......)))). ( -23.30) >DroEre_CAF1 25089 80 + 1 CU--------------------------------UAUUUGGCCAGGACUUGCCAGUCAUUUUGGCCAGGACGACACCCUCGUCUUGG-AUUUUAUUUGCCACCUUUCUGGGCC ..--------------------------------.....((((.(((...(((((.....)))))((((((((.....)))))))).-.................))).)))) ( -25.10) >DroYak_CAF1 26501 113 + 1 CAGCGCCAUCCUUAUCCUUUAUCUUUUUUUUUUGUUUUUGGCCAGAACUUGCCAGCCAUUUUGGCCAGGAGAACACCCUCGUCUUGAAAUUUUAUUUGCCACCUUUCUGGGCC ....(((..............................((((((((((...(....)..))))))))))(((......))).............................))). ( -19.10) >consensus CAGCCCCAUCCAUAUUUU_UUUC__________UUUUUUGGCCAAAACUUGCCAGCCAUUUUGGCCAGGACAACACCCUCGUCUUGGAAUUUUAUUUGCCACCUUUCUGGGCC .......................................((((.......(((((.....)))))((((((.........)))))).......................)))) (-17.30 = -17.50 + 0.20)

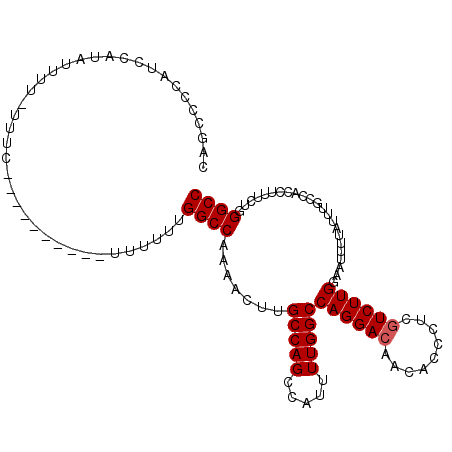
| Location | 11,986,922 – 11,987,023 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11986922 101 - 22224390 GGCCCAGAAAGGUGGCAAAUAAAAUUCCAAGACGAGGGUGUUGUCCUGGCCAAAAUGGCUGGCAAGUUUUGGCCAAAAAA-----------AAA-AAAAAAAGGAUGGGGCUG (((((.....((.(((((...(..(((......)))..).)))))))(((((((((.........)))))))))......-----------...-............))))). ( -27.10) >DroSec_CAF1 26308 106 - 1 GGCCCAGAAAGGUGGCAAAUAAAAUUCCAAGACGAGGGUGUUGUCCUGGCCAAAAUGGCUGGCAAGUUUUGGCCAAAAAAGGGC------GAAA-AAAAUAUGGAUGGGGCUC (((((......((.....)).....((((..((....)).((((((((((((((((.........))))))))).....)))))------))..-......))))..))))). ( -28.20) >DroSim_CAF1 26914 107 - 1 GGCCCAGAAAGGUGGCAAAUAAAAUUCCAAGACGAGGGUGUUGUCCUGGCCAAAAUGGCUGGCAAGUUUUGGCCAAAAAAGGGC------GAAAAAAAAUAUGGAUGGGGCUG (((((......((.....)).....((((...........((((((((((((((((.........))))))))).....)))))------)).........))))..))))). ( -28.95) >DroEre_CAF1 25089 80 - 1 GGCCCAGAAAGGUGGCAAAUAAAAU-CCAAGACGAGGGUGUCGUCCUGGCCAAAAUGACUGGCAAGUCCUGGCCAAAUA--------------------------------AG .((((......).))).........-....(((((.....))))).((((((....((((....)))).))))))....--------------------------------.. ( -22.00) >DroYak_CAF1 26501 113 - 1 GGCCCAGAAAGGUGGCAAAUAAAAUUUCAAGACGAGGGUGUUCUCCUGGCCAAAAUGGCUGGCAAGUUCUGGCCAAAAACAAAAAAAAAAGAUAAAGGAUAAGGAUGGCGCUG .((((......).)))....................(((((..((((((((.....))))(((........)))...........................))))..))))). ( -25.60) >consensus GGCCCAGAAAGGUGGCAAAUAAAAUUCCAAGACGAGGGUGUUGUCCUGGCCAAAAUGGCUGGCAAGUUUUGGCCAAAAAA__________GAAA_AAAAUAAGGAUGGGGCUG ((((.((((...((.((..........((.(((((.....))))).))(((.....))))).))..))))))))....................................... (-20.50 = -20.70 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:08 2006