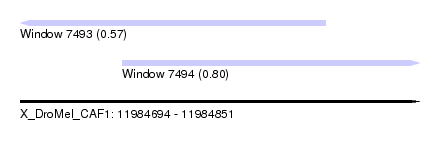
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,984,694 – 11,984,851 |
| Length | 157 |
| Max. P | 0.797342 |

| Location | 11,984,694 – 11,984,814 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.55 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -19.47 |
| Energy contribution | -19.22 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
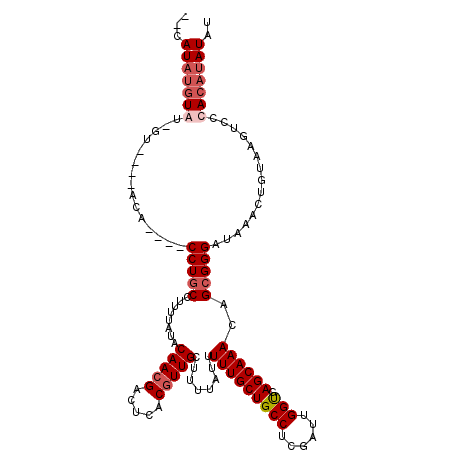
>X_DroMel_CAF1 11984694 120 - 22224390 CACCAAUCGAGGCAGCAAAAAUAAAAGCAACGUGAGUCGUUGUAUAAAAGGCAGGCAGGUGUAUGUACAAUACAUAUGUACAUACAUACGUACGCUGGCACACGCGGCGUAUGCGCAAUU ...........((.((.........(((.((....)).))).........))..((..(((((((((((.......)))))))))))..((((((((.......)))))))))))).... ( -35.37) >DroSec_CAF1 24100 103 - 1 CACCAAUCGAGGCAGCAAAAAUAAAAGCAACGUGAGUCGUUGUAUAAAAGGCAGG----UGU----AC-AUACAUAUG--------UACGUACGCUGGCACACGCGGCGUAUGCGCAAUU ..((......))..((..........((((((.....)))))).......((.((----(((----((-.(((....)--------)).))))))).))....)).(((....))).... ( -27.90) >DroSim_CAF1 24707 103 - 1 CACCAAUCGAGGCAGCAAAAAUAAAAGCAACGUGAGUCGUUGUAUAAAAGGCAGG----UGU----AC-AUACAUAUG--------UACGUACGCUGGCACACGCGGCGUAUGCGCAAUU ..((......))..((..........((((((.....)))))).......((.((----(((----((-.(((....)--------)).))))))).))....)).(((....))).... ( -27.90) >DroYak_CAF1 24369 103 - 1 CGCCAAUCGAGGCAGCAAAAAUAAAAGCAACGUGAGUCCUUGUAUAAAAGGUAGG----UAU----AU-GUAUGUAUG--------UUUAAAUGCUGGCACACGCGGCGUAUGCGCAAUU .(((......))).((....(((...((..((((.(((...((((.(((.(((.(----((.----..-.))).))).--------)))..)))).))).))))..)).)))..)).... ( -24.80) >consensus CACCAAUCGAGGCAGCAAAAAUAAAAGCAACGUGAGUCGUUGUAUAAAAGGCAGG____UGU____AC_AUACAUAUG________UACGUACGCUGGCACACGCGGCGUAUGCGCAAUU ...........((.((.........(((.((....)).))).........))....................................(((((((((.......))))))))).)).... (-19.47 = -19.22 + -0.25)



| Location | 11,984,734 – 11,984,851 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.54 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -19.62 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11984734 117 + 22224390 UACAUAUGUAUUGUACAUACACCUGCCUGCCUUUUAUACAACGACUCACGUUGCUUUUAUUUUUGCUGCCUCGAUUGGUGAGCAAACAGCGGGAUAAACUGUAAGUCCUACAUAUAU ...(((((((..(.((.((((....(((((........(((((.....)))))........(((((((((......))).))))))..)))))......)))).)).)))))))).. ( -28.60) >DroSec_CAF1 24134 106 + 1 --CAUAUGUAU-GU----ACA----CCUGCCUUUUAUACAACGACUCACGUUGCUUUUAUUUUUGCUGCCUCGAUUGGUGAGCAAACAGCGGGAUAAACUGUAAGUCUCACAUAUAU --...((((((-((----...----(((((........(((((.....)))))........(((((((((......))).))))))..)))))....((.....))...)))))))) ( -25.40) >DroSim_CAF1 24741 106 + 1 --CAUAUGUAU-GU----ACA----CCUGCCUUUUAUACAACGACUCACGUUGCUUUUAUUUUUGCUGCCUCGAUUGGUGAGCAAACAGCGGGAUAAACUGUAAGUCCCACAUAUAU --...((((((-((----...----...((........(((((.....)))))........(((((((((......))).))))))..))(((((.........))))))))))))) ( -27.00) >DroYak_CAF1 24403 106 + 1 --CAUACAUAC-AU----AUA----CCUACCUUUUAUACAAGGACUCACGUUGCUUUUAUUUUUGCUGCCUCGAUUGGCGAGCAAACAGCGGGAUAAACUGAAAGUCCUACACAUAU --.........-..----...----...............((((((((.(((..(((..(((((((((((......))).)))))).))..)))..))))))..)))))........ ( -21.50) >consensus __CAUAUGUAU_GU____ACA____CCUGCCUUUUAUACAACGACUCACGUUGCUUUUAUUUUUGCUGCCUCGAUUGGUGAGCAAACAGCGGGAUAAACUGUAAGUCCCACAUAUAU ...(((((((...............(((((........(((((.....)))))........(((((((((......))).))))))..)))))...............))))))).. (-19.62 = -21.18 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:05 2006