
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,981,347 – 11,981,458 |
| Length | 111 |
| Max. P | 0.904702 |

| Location | 11,981,347 – 11,981,458 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -39.94 |
| Consensus MFE | -32.44 |
| Energy contribution | -32.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
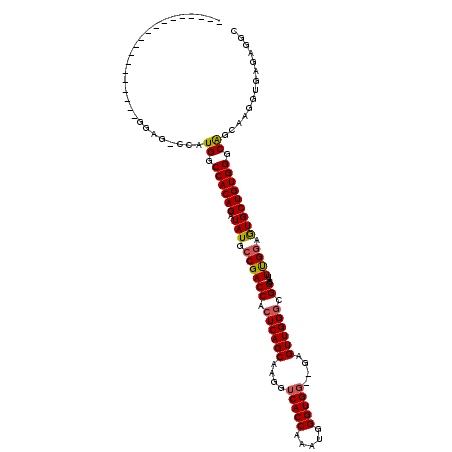
>X_DroMel_CAF1 11981347 111 + 22224390 GUCGUUCCACU--------GGAG-CCAUGGCCACAGAUAUGACGACCACUCAGCAAGGUCACCAAAUGGGUGGGGGAGUUGGGCGGCAUUUGGAGUGCUGUGGGCAGCAAGGUGAGAGGC (((.(..((((--------...(-(..((.((((((.(((..(((((.((((((....(((((.....)))))....)))))).))...)))..))))))))).))))..))))..)))) ( -39.30) >DroSec_CAF1 21108 98 + 1 -------------------GGAG-ACAUGGCCACAGAUAUGCCGACCACUCAGCAAGGUCACCAAAUGGGUGG--GAGUUGGGCGGCACUCGGAGUGCUGUGGGCAGCAAGGUGAGAGGC -------------------(...-.)...(((.......(((((.((((((.......(((((.....)))))--))).))).)))))((((...(((((....)))))...)))).))) ( -36.21) >DroSim_CAF1 21166 98 + 1 -------------------GGAG-CCAUGGCCACAGAUAUGCCGACCACUCAGCAAGGUCACCAAAUGGGUGG--GAGUUGGGCGGCAUUCGGAGUGCUGUGGGCAGCAAGGUGAGAGGC -------------------...(-((.((.((((((.(((.((((((.((((((....(((((.....)))))--..)))))).))...)))).))))))))).))....)))....... ( -37.50) >DroEre_CAF1 19792 99 + 1 -----------------GUGGAA-CCCUGGCCACAGAUAUGCCGACCACUCAGCAAGGUCACCA-AUGGGUGC--GAGUUGGUCGGCAUUUGGAGUGCUGUGGGCGGCAAGGUGAGAGGC -----------------......-((((.(((.((((..((((((((((((.(((...(((...-.))).)))--))).)))))))))))))...(((((....))))).))).)).)). ( -40.70) >DroYak_CAF1 20790 115 + 1 G---UUCCACUGCAGCCAUGGAAGCCAUGGCCACAGAUAUGCCGACCACUCAGCAAGGUCACCAAAUGGGUGG--GAGUUGGGCGGCAUUUGGAAUGCUGUGGGCAGCAAGGUGAGAGGC (---((((((((..(((((((...)))))))..)))..((((((.((((((.......(((((.....)))))--))).))).)))))).))))))((((....))))............ ( -46.01) >consensus ___________________GGAG_CCAUGGCCACAGAUAUGCCGACCACUCAGCAAGGUCACCAAAUGGGUGG__GAGUUGGGCGGCAUUUGGAGUGCUGUGGGCAGCAAGGUGAGAGGC ...........................((.((((((.(((.((((((.((((((....(((((.....)))))....)))))).))...)))).))))))))).)).............. (-32.44 = -32.48 + 0.04)

| Location | 11,981,347 – 11,981,458 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
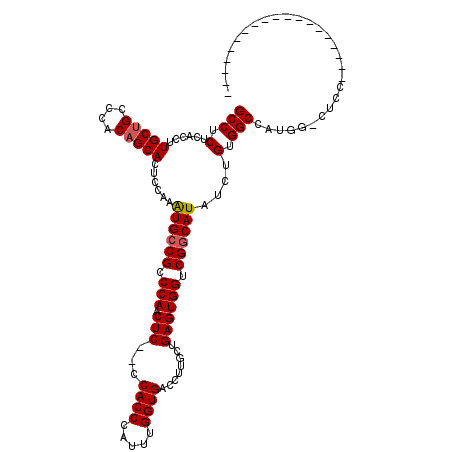
>X_DroMel_CAF1 11981347 111 - 22224390 GCCUCUCACCUUGCUGCCCACAGCACUCCAAAUGCCGCCCAACUCCCCCACCCAUUUGGUGACCUUGCUGAGUGGUCGUCAUAUCUGUGGCCAUGG-CUCC--------AGUGGAACGAC .((.((((.(.(((((....)))))(.(((((((..................))))))).).....).)))).))((((..((.(((..((....)-)..)--------))))..)))). ( -29.77) >DroSec_CAF1 21108 98 - 1 GCCUCUCACCUUGCUGCCCACAGCACUCCGAGUGCCGCCCAACUC--CCACCCAUUUGGUGACCUUGCUGAGUGGUCGGCAUAUCUGUGGCCAUGU-CUCC------------------- (((.(......((((((((.(((((....((((........))))--.((((.....))))....))))).).)).))))).....).))).....-....------------------- ( -29.50) >DroSim_CAF1 21166 98 - 1 GCCUCUCACCUUGCUGCCCACAGCACUCCGAAUGCCGCCCAACUC--CCACCCAUUUGGUGACCUUGCUGAGUGGUCGGCAUAUCUGUGGCCAUGG-CUCC------------------- (((..((((..(((((....)))))....((((((((.(((.(((--.((((.....))))........)))))).)))))).)).))))....))-)...------------------- ( -30.90) >DroEre_CAF1 19792 99 - 1 GCCUCUCACCUUGCCGCCCACAGCACUCCAAAUGCCGACCAACUC--GCACCCAU-UGGUGACCUUGCUGAGUGGUCGGCAUAUCUGUGGCCAGGG-UUCCAC----------------- .......((((((..(((.((((........((((((((((.(((--(((((...-.)))).......))))))))))))))..))))))))))))-).....----------------- ( -36.91) >DroYak_CAF1 20790 115 - 1 GCCUCUCACCUUGCUGCCCACAGCAUUCCAAAUGCCGCCCAACUC--CCACCCAUUUGGUGACCUUGCUGAGUGGUCGGCAUAUCUGUGGCCAUGGCUUCCAUGGCUGCAGUGGAA---C ...........(((((....)))))(((((.((((((.(((.(((--.((((.....))))........)))))).))))))..(((..(((((((...)))))))..))))))))---. ( -47.40) >consensus GCCUCUCACCUUGCUGCCCACAGCACUCCAAAUGCCGCCCAACUC__CCACCCAUUUGGUGACCUUGCUGAGUGGUCGGCAUAUCUGUGGCCAUGG_CUCC___________________ (((.(......(((((....)))))......((((((.(((.(((...((((.....))))........)))))).))))))....).)))............................. (-26.08 = -26.32 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:04 2006