
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,980,205 – 11,980,313 |
| Length | 108 |
| Max. P | 0.785375 |

| Location | 11,980,205 – 11,980,313 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -10.50 |
| Energy contribution | -12.92 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
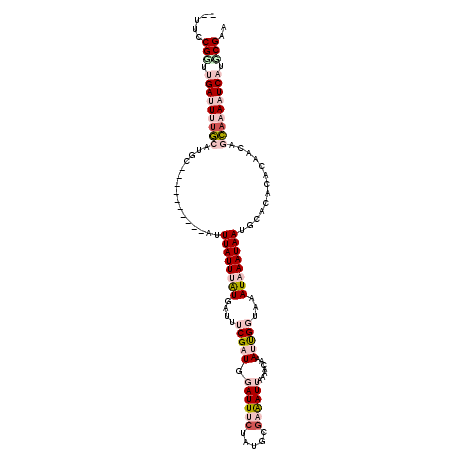
>X_DroMel_CAF1 11980205 108 + 22224390 --UUCCGGUUGAUUUUGCAUGC----------AUUUAUUUAUGAUUUCGAUGGAUUUCUAUGGGAAAUUAAAACAAAUUGGUAAAUAAAUAAUGCACACACAACAGCAAAAUCAUGCGAA --...((..(((((((((.(((----------(((.((((((....(((((.((((((.....)))))).......)))))...)))))))))))).........)))))))))..)).. ( -23.40) >DroPse_CAF1 116540 116 + 1 ACUCCCGUUCGAAUGUCCAUUCCGGCACUCCAAUUUAUUCAUGAUUCCGAUGGAUUACUUUGCGAGAUGCAAACAUAACGCAAAAUAAAUAAUACACAAGC----UGAAAAUCAUGCGAA .......((((.(((.......((((...........(((((.......)))))(((.((((((..(((....)))..)))))).)))...........))----)).....))).)))) ( -18.30) >DroSec_CAF1 19993 108 + 1 --UUCCGGUUGAUUUUGCAUGC----------AUUUAUUUGUGAUUUCGAUGGAUUUCUAUGGGAAAUUAAAACAAAUUGGUUAAUAAAUAAUGCACACACAACGGCAAAAUCAUGCGAA --...((..(((((((((.(((----------(((.((((((..((((.((((....)))).)))).....(((......))).))))))))))))..(.....))))))))))..)).. ( -24.90) >DroEre_CAF1 18782 108 + 1 --UUUCGCAUGAUUUCGCAUGC----------AUUUAUUUAUGAUUUCGAUGGAUUUCUAUGGUAAAUUAAAAUAAAUUGGUAAAUAAAUAAUGCACACACAACAGCAAAAUCAUGCGAA --.((((((((((((.((.(((----------(((.((((((....(((((..((((..((.....))..))))..)))))...)))))))))))).........)).)))))))))))) ( -28.50) >DroYak_CAF1 19664 108 + 1 --UUCCGGAUGAUUUUGCAUGC----------AUUUAUUUAUGAUUUCGAUGGAUUUCUAUGCGAAAUUAAAACAAAUUGGUAAAUAAAUAAUGCACAUGCAACAGCAAAAUCAUUCGAA --...(((((((((((((....----------(((((((((((((((((((((....)))).)))))))....(.....)))))))))))..(((....)))...))))))))))))).. ( -34.10) >DroPer_CAF1 121391 116 + 1 ACUCCCGUUCGAAUGUCCAUUCCGGCACUCCAAUUUAUUCAUGAUUCCGAUGGAUUACUUUGCGAGAUGCAAACAUAACGCAAAAUAAAUAAUACACAAGC----UGAAAAUCAUGCGAA .......((((.(((.......((((...........(((((.......)))))(((.((((((..(((....)))..)))))).)))...........))----)).....))).)))) ( -18.30) >consensus __UUCCGGUUGAUUUUGCAUGC__________AUUUAUUUAUGAUUUCGAUGGAUUUCUAUGCGAAAUUAAAACAAAUUGGUAAAUAAAUAAUGCACACACAACAGCAAAAUCAUGCGAA .....(((.(((((((((................((((((((....(((((.((((((.....)))))).......)))))...)))))))).............))))))))).))).. (-10.50 = -12.92 + 2.42)

| Location | 11,980,205 – 11,980,313 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -11.69 |
| Energy contribution | -13.28 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11980205 108 - 22224390 UUCGCAUGAUUUUGCUGUUGUGUGUGCAUUAUUUAUUUACCAAUUUGUUUUAAUUUCCCAUAGAAAUCCAUCGAAAUCAUAAAUAAAU----------GCAUGCAAAAUCAACCGGAA-- ((((..(((((((((........(((((((...((((((....((((.....(((((.....)))))....))))....)))))))))----------)))))))))))))..)))).-- ( -21.60) >DroPse_CAF1 116540 116 - 1 UUCGCAUGAUUUUCA----GCUUGUGUAUUAUUUAUUUUGCGUUAUGUUUGCAUCUCGCAAAGUAAUCCAUCGGAAUCAUGAAUAAAUUGGAGUGCCGGAAUGGACAUUCGAACGGGAGU (((((((((((((..----((....)).....((((((((((..(((....)))..))))))))))......)))))))))......((.(((((((.....)).))))).))))))... ( -28.80) >DroSec_CAF1 19993 108 - 1 UUCGCAUGAUUUUGCCGUUGUGUGUGCAUUAUUUAUUAACCAAUUUGUUUUAAUUUCCCAUAGAAAUCCAUCGAAAUCACAAAUAAAU----------GCAUGCAAAAUCAACCGGAA-- ((((..(((((((((........(((((((...((((......((((.....(((((.....)))))....))))......)))))))----------)))))))))))))..)))).-- ( -19.00) >DroEre_CAF1 18782 108 - 1 UUCGCAUGAUUUUGCUGUUGUGUGUGCAUUAUUUAUUUACCAAUUUAUUUUAAUUUACCAUAGAAAUCCAUCGAAAUCAUAAAUAAAU----------GCAUGCGAAAUCAUGCGAAA-- (((((((((((((((........(((((((((((((...........(((((........))))).......(....))))))).)))----------))))))))))))))))))).-- ( -27.90) >DroYak_CAF1 19664 108 - 1 UUCGAAUGAUUUUGCUGUUGCAUGUGCAUUAUUUAUUUACCAAUUUGUUUUAAUUUCGCAUAGAAAUCCAUCGAAAUCAUAAAUAAAU----------GCAUGCAAAAUCAUCCGGAA-- ((((.((((((((((((((((....)))..(((((((((....((((.....(((((.....)))))....))))....)))))))))----------))).)))))))))).)))).-- ( -27.10) >DroPer_CAF1 121391 116 - 1 UUCGCAUGAUUUUCA----GCUUGUGUAUUAUUUAUUUUGCGUUAUGUUUGCAUCUCGCAAAGUAAUCCAUCGGAAUCAUGAAUAAAUUGGAGUGCCGGAAUGGACAUUCGAACGGGAGU (((((((((((((..----((....)).....((((((((((..(((....)))..))))))))))......)))))))))......((.(((((((.....)).))))).))))))... ( -28.80) >consensus UUCGCAUGAUUUUGCUGUUGCGUGUGCAUUAUUUAUUUACCAAUUUGUUUUAAUUUCCCAUAGAAAUCCAUCGAAAUCAUAAAUAAAU__________GCAUGCAAAAUCAACCGGAA__ ((((..(((((((((...(((.........((((((((.....((((.....(((((.....)))))....)))).....))))))))..........))).)))))))))..))))... (-11.69 = -13.28 + 1.59)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:02 2006