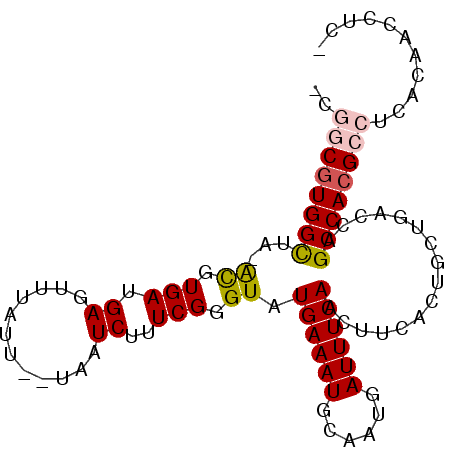
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,339,362 – 1,339,457 |
| Length | 95 |
| Max. P | 0.892290 |

| Location | 1,339,362 – 1,339,457 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -18.19 |
| Energy contribution | -19.42 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1339362 95 + 22224390 -GAGGCGUUGAGGCGUGGCUGGUCAGCAGUGAAGUUGAAAUCAUUGAAUUUCAUACCCGAAAGAUUAGUAAUAAACUCAUCACGC-UAGCCACGCCG- -..........((((((((((((.....((((((((.((....)).))))))))........(((.(((.....))).)))..))-)))))))))).- ( -34.10) >DroPse_CAF1 3779 92 + 1 UGGGCUUGUGAGGCGUGGCAUAUCAGCU-UUAAGUUGAAAUCAUUGCAUUUCAUACCCGAAAGAU----AAAAAAAUCAUCAUAU-GAACCACGGGGU ......(((((((.(..(.((.(((((.-....))))).))..)..).)))))))((((...(((----......))).((....-))....)))).. ( -17.90) >DroSim_CAF1 3082 95 + 1 -GAGGCGUUGAGGCGUGGCUGGUCAGCAGUGAAGUUGAAAUCAUUGAAUUUCAUACCCGAAAGAUUAGUAAUAAACUCAUCACGC-UAGCCACGCCG- -..........((((((((((((.....((((((((.((....)).))))))))........(((.(((.....))).)))..))-)))))))))).- ( -34.10) >DroYak_CAF1 70 95 + 1 -GAGGCAGUGAGGCGUGGCUGGUCAGCAGUGAAGUUGAAAUCAUUGAAUUUCAUACCCGAAAGAUUAGUGAUAAACUCAUCACGU-UGGCCACGCCG- -..........((((((((....((((.((((((((...((((((((.((((......))))..)))))))).))))..))))))-)))))))))).- ( -35.00) >DroAna_CAF1 4431 93 + 1 -UCCAUAGUGAGGCUUGGCUGGUCAGCGGUGAAAGUGAAAUCAUUGCAUUUCAUACCCGAAAGAUUA---AAAGAAUCAUCACGUGCAGCCACGCCU- -.........((((.((((((....(.(((((..(((((((......)))))))...(....)....---......))))).)...)))))).))))- ( -27.80) >DroPer_CAF1 3765 92 + 1 UGGGCUUGUGAGGCGUGGCAUAUCAGCU-UUAAGUUGAAAUCAUUGCAUUUCAUACCCGAAAGAU----ACAAAAAUCAUCAUAU-GAACCACGGGGU ......(((((((.(..(.((.(((((.-....))))).))..)..).)))))))((((...(((----......))).((....-))....)))).. ( -17.90) >consensus _GAGGCAGUGAGGCGUGGCUGGUCAGCAGUGAAGUUGAAAUCAUUGAAUUUCAUACCCGAAAGAUUA__AAAAAAAUCAUCACGU_UAGCCACGCCG_ ...........((((((((((.(((((......)))))...................(....).......................)))))))))).. (-18.19 = -19.42 + 1.22)

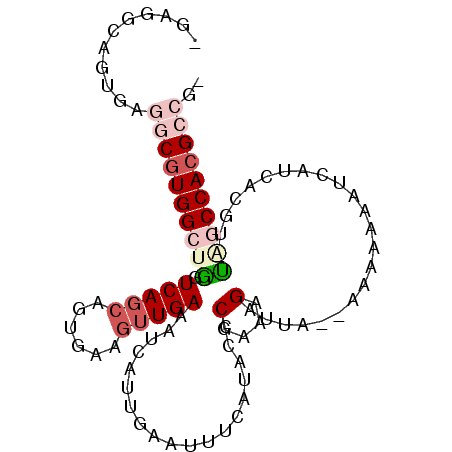
| Location | 1,339,362 – 1,339,457 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1339362 95 - 22224390 -CGGCGUGGCUA-GCGUGAUGAGUUUAUUACUAAUCUUUCGGGUAUGAAAUUCAAUGAUUUCAACUUCACUGCUGACCAGCCACGCCUCAACGCCUC- -.((((((((((-(((((((((((((..((((.........))))..))))))).((....))...)))).)))....)))))))))..........- ( -31.10) >DroPse_CAF1 3779 92 - 1 ACCCCGUGGUUC-AUAUGAUGAUUUUUUU----AUCUUUCGGGUAUGAAAUGCAAUGAUUUCAACUUAA-AGCUGAUAUGCCACGCCUCACAAGCCCA ....(((((..(-(((((((((.....))----)))(((..(((.((((((......)))))))))..)-))...))))))))))............. ( -18.20) >DroSim_CAF1 3082 95 - 1 -CGGCGUGGCUA-GCGUGAUGAGUUUAUUACUAAUCUUUCGGGUAUGAAAUUCAAUGAUUUCAACUUCACUGCUGACCAGCCACGCCUCAACGCCUC- -.((((((((((-(((((((((((((..((((.........))))..))))))).((....))...)))).)))....)))))))))..........- ( -31.10) >DroYak_CAF1 70 95 - 1 -CGGCGUGGCCA-ACGUGAUGAGUUUAUCACUAAUCUUUCGGGUAUGAAAUUCAAUGAUUUCAACUUCACUGCUGACCAGCCACGCCUCACUGCCUC- -.((((((((..-..((((((....)))))).........((...(((((((....)))))))...(((....))))).))))))))..........- ( -28.50) >DroAna_CAF1 4431 93 - 1 -AGGCGUGGCUGCACGUGAUGAUUCUUU---UAAUCUUUCGGGUAUGAAAUGCAAUGAUUUCACUUUCACCGCUGACCAGCCAAGCCUCACUAUGGA- -((((.((((((((.(((..((((....---.))))....(((..((((((......))))))..)))..)))))..)))))).)))).........- ( -23.90) >DroPer_CAF1 3765 92 - 1 ACCCCGUGGUUC-AUAUGAUGAUUUUUGU----AUCUUUCGGGUAUGAAAUGCAAUGAUUUCAACUUAA-AGCUGAUAUGCCACGCCUCACAAGCCCA ....((((((((-((...)))).....((----((((((..(((.((((((......)))))))))..)-))..)))))))))))............. ( -17.50) >consensus _CGGCGUGGCUA_ACGUGAUGAGUUUAUU__UAAUCUUUCGGGUAUGAAAUGCAAUGAUUUCAACUUCACUGCUGACCAGCCACGCCUCACAACCUC_ ..((((((((...((.(((.((............))..))).)).((((((......))))))................))))))))........... (-17.38 = -17.55 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:57 2006