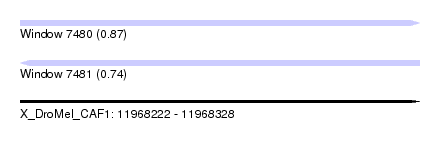
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,968,222 – 11,968,328 |
| Length | 106 |
| Max. P | 0.872594 |

| Location | 11,968,222 – 11,968,328 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.19 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -24.96 |
| Energy contribution | -26.90 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11968222 106 + 22224390 CAGCACGCCAGGGUAAUUUAUCGAUAGCAUUGGGCAUAUCGAUUACUGUGGCUGCAGCGUCAGCUAAAUUGAUUGCUUGCACA----------ACUGUACUAAGAAGUGCAUCAAU ..(((.((((.((((....(((((((((.....)).))))))))))).)))))))(((....)))......((((..(((((.----------.((......))..))))).)))) ( -36.00) >DroSec_CAF1 7584 106 + 1 CAGCACGCCAGGGUAAUUUAUCGAUAGCAUUGGGCAUAUCGAUUACUGUGGCUGCAGUGUCAUCUAAAUUGAUUCCUAGCACA----------ACUGUACUAAGAAGUGCAUCAAU ..(((.((((.((((....(((((((((.....)).))))))))))).)))))))((.((((.......))))..)).((((.----------.((......))..))))...... ( -32.50) >DroSim_CAF1 7640 106 + 1 CAGCGCGCCAGGGUAAUUUAUCGAUAGCAUUGGGCAUAUCGAUUACUGGGGCUGCAGUGUCAGCUAAAUUGAUUGCUAGCACA----------ACUGUACUAAGAAGUGCAUCAAU ..(((.(((..((((....(((((((((.....)).)))))))))))..)))))).(((..(((..........)))..))).----------..((((((....))))))..... ( -33.70) >DroEre_CAF1 7883 116 + 1 CAGCACGCCAUGGAAAUUUAUCGAUAGCAUGGGGCAUAUCGAUUAUUGCGGCUUGAGUGCCAGUUAAGUUGAUUCCUCGCACAAACUUGCGUAACUGUACUGAGGAGGGCAUCGAU ......(((.....((((((((((((((.....)).)))))).......(((......)))...))))))..(((((((((......)))(((....))).)))))))))...... ( -30.40) >consensus CAGCACGCCAGGGUAAUUUAUCGAUAGCAUUGGGCAUAUCGAUUACUGUGGCUGCAGUGUCAGCUAAAUUGAUUCCUAGCACA__________ACUGUACUAAGAAGUGCAUCAAU ..(((.((((.((((....(((((((((.....)).))))))))))).))))))).((((.(((..........))).)))).............((((((....))))))..... (-24.96 = -26.90 + 1.94)


| Location | 11,968,222 – 11,968,328 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.19 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -19.88 |
| Energy contribution | -22.50 |
| Covariance contribution | 2.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11968222 106 - 22224390 AUUGAUGCACUUCUUAGUACAGU----------UGUGCAAGCAAUCAAUUUAGCUGACGCUGCAGCCACAGUAAUCGAUAUGCCCAAUGCUAUCGAUAAAUUACCCUGGCGUGCUG .....(((((..((......)).----------.)))))((((.......((((....))))..((((..((((((((((.((.....)))))))))....)))..)))).)))). ( -28.70) >DroSec_CAF1 7584 106 - 1 AUUGAUGCACUUCUUAGUACAGU----------UGUGCUAGGAAUCAAUUUAGAUGACACUGCAGCCACAGUAAUCGAUAUGCCCAAUGCUAUCGAUAAAUUACCCUGGCGUGCUG ......((((((((((((((...----------.))))))))))....................((((..((((((((((.((.....)))))))))....)))..)))))))).. ( -32.90) >DroSim_CAF1 7640 106 - 1 AUUGAUGCACUUCUUAGUACAGU----------UGUGCUAGCAAUCAAUUUAGCUGACACUGCAGCCCCAGUAAUCGAUAUGCCCAAUGCUAUCGAUAAAUUACCCUGGCGCGCUG ...................((((----------.(((((((...........((((......)))).......(((((((.((.....)))))))))........))))))))))) ( -26.00) >DroEre_CAF1 7883 116 - 1 AUCGAUGCCCUCCUCAGUACAGUUACGCAAGUUUGUGCGAGGAAUCAACUUAACUGGCACUCAAGCCGCAAUAAUCGAUAUGCCCCAUGCUAUCGAUAAAUUUCCAUGGCGUGCUG ((((((((..(((((.((((((..((....)))))))))))))............(((......))))))....)))))......((((((((.((......)).))))))))... ( -36.20) >consensus AUUGAUGCACUUCUUAGUACAGU__________UGUGCUAGCAAUCAAUUUAGCUGACACUGCAGCCACAGUAAUCGAUAUGCCCAAUGCUAUCGAUAAAUUACCCUGGCGUGCUG ......(((((((((((((((............)))))))))))....................((((..((((((((((.((.....)))))))))....)))..)))))))).. (-19.88 = -22.50 + 2.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:53 2006