
| Sequence ID | X_DroMel_CAF1 |
|---|---|
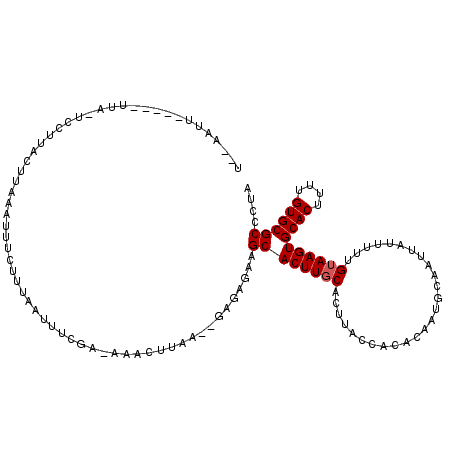
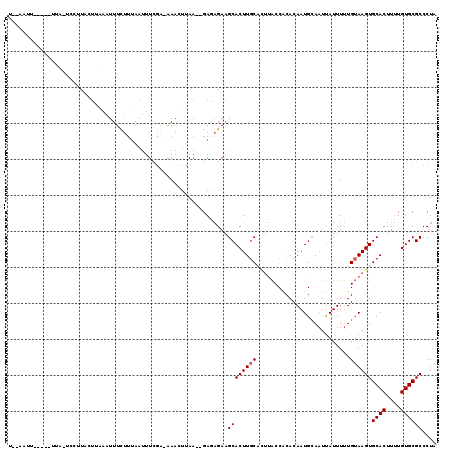
| Location | 11,961,502 – 11,961,610 |
| Length | 108 |
| Max. P | 0.965401 |

| Location | 11,961,502 – 11,961,610 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -9.99 |
| Energy contribution | -10.24 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11961502 108 + 22224390 U--AAUU-----UAA-UUCUUCCUUUAAUUUCUUGAAUUUCGA-AAACUUAA--GAGAGAAGCACUUGCACUUACCGCACAAUGCAAUCAUUUUUGUAAGUGCACUUUUGUGCGCCCUA .--....-----...-((((..(((...((((.........))-))....))--)..))))((((.(((((((((.....((((....))))...))))))))).....))))...... ( -22.70) >DroSec_CAF1 826 109 + 1 C--AAUU-----UUA-UCCUUUUUUAAAUUUCUUUAAUUUCGGAAAACUUAA--GAAAGAAGCACUUGCUUUUACCACACAAUGCAAUUAUUUUUGUAAGUGCACUUUUGUGCGCCCUA .--....-----...-..(((((((((.(((((........)))))..))))--)))))((((....))))...........(((((......))))).((((((....)))))).... ( -21.80) >DroSim_CAF1 818 115 + 1 UUAAAUUAGAAAUUAUUCCUAAUUU---UUUUUUUAAAUUUGA-GAAAUUAAGAGACAGAAGCACUUGCACUUACCACACAAUGAAAUUAUUUUUGUAAGUGCACUUUUGUGCGCCCUA (((((..((((((((....))))))---))..))))).(((((-....)))))..(((((((....(((((((((.....((((....))))...))))))))))))))))........ ( -22.80) >DroEre_CAF1 1361 108 + 1 U--CAUU-UAAUUUA-----UACCUAAAUUUCUAUUAUUUUUA-GAAGCUAA--GAGUGAUGCACUUACUGCCGCCACACAAUGCACUUAUUUCUGAAAGUGCACUUCUGUGCGCCCUA .--....-.......-----.............((((((((((-(...))))--)))))))(((.....))).((((((.(((((((((........))))))).)).)))).)).... ( -21.80) >consensus U__AAUU_____UUA_UCCUUACUUAAAUUUCUUUAAUUUCGA_AAACUUAA__GAGAGAAGCACUUGCACUUACCACACAAUGCAAUUAUUUUUGUAAGUGCACUUUUGUGCGCCCUA .............................................................((((((((..........................))))))((((....)))))).... ( -9.99 = -10.24 + 0.25)

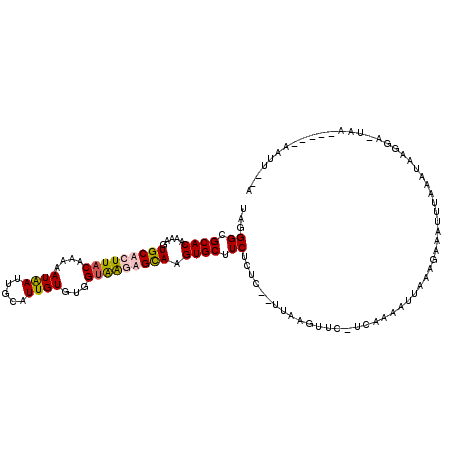
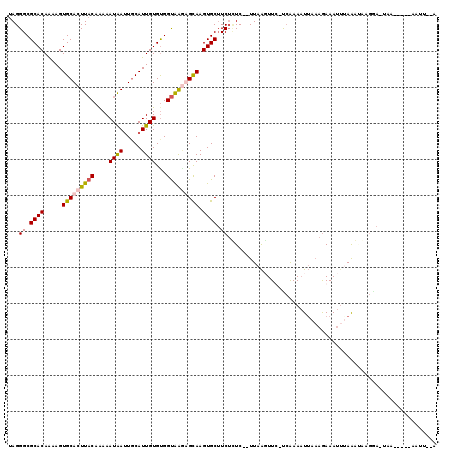
| Location | 11,961,502 – 11,961,610 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -13.73 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11961502 108 - 22224390 UAGGGCGCACAAAAGUGCACUUACAAAAAUGAUUGCAUUGUGCGGUAAGUGCAAGUGCUUCUCUC--UUAAGUUU-UCGAAAUUCAAGAAAUUAAAGGAAGAA-UUA-----AAUU--A ......((((.....(((((((((...((((....)))).....))))))))).))))((((..(--((.(((((-(..........)))))).)))..))))-...-----....--. ( -26.70) >DroSec_CAF1 826 109 - 1 UAGGGCGCACAAAAGUGCACUUACAAAAAUAAUUGCAUUGUGUGGUAAAAGCAAGUGCUUCUUUC--UUAAGUUUUCCGAAAUUAAAGAAAUUUAAAAAAGGA-UAA-----AAUU--G (((((((((((...(((((.(((......))).)))))))))))....((((....))))...))--)))((((((((....(((((....)))))....)))-..)-----))))--. ( -21.50) >DroSim_CAF1 818 115 - 1 UAGGGCGCACAAAAGUGCACUUACAAAAAUAAUUUCAUUGUGUGGUAAGUGCAAGUGCUUCUGUCUCUUAAUUUC-UCAAAUUUAAAAAAA---AAAUUAGGAAUAAUUUCUAAUUUAA (((((.((((.....(((((((((....(((((...)))))...))))))))).)))))))))............-...............---(((((((((.....))))))))).. ( -26.10) >DroEre_CAF1 1361 108 - 1 UAGGGCGCACAGAAGUGCACUUUCAGAAAUAAGUGCAUUGUGUGGCGGCAGUAAGUGCAUCACUC--UUAGCUUC-UAAAAAUAAUAGAAAUUUAGGUA-----UAAAUUA-AAUG--A ...(.((((((...((((((((........)))))))))))))).)(((.(..((((...)))).--.).)))((-((.......))))((((((....-----)))))).-....--. ( -24.90) >consensus UAGGGCGCACAAAAGUGCACUUACAAAAAUAAUUGCAUUGUGUGGUAAGAGCAAGUGCUUCUCUC__UUAAGUUC_UCAAAAUUAAAGAAAUUUAAAUAAGGA_UAA_____AAUU__A ...((.((((.....(((((((((....((((.....))))...))))))))).)))).)).......................................................... (-13.73 = -14.22 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:48 2006