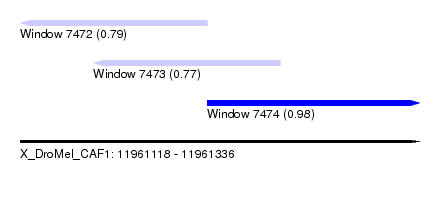
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,961,118 – 11,961,336 |
| Length | 218 |
| Max. P | 0.979494 |

| Location | 11,961,118 – 11,961,220 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11961118 102 - 22224390 AAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCUUAGUGGA---UGGCGUGGGUGGCGCCAGUGGAAAGUGCUGGGGUAAUGGGA ......((((((.(((.((.(((..(......)....(((((((....)))))))..))).---)).))).)))))).(((((.......))))).......... ( -35.00) >DroSec_CAF1 405 101 - 1 AAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCGUAGUGGA---UGGCGUGGGUGGCGC-AAUGGAAAGUGCUGGGGUAAUGGGA ......((((((.(((.((.(((..(......)......(((((....)))))....))).---)).))).))))))((-(.(....).)))............. ( -29.70) >DroSim_CAF1 406 101 - 1 AAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCGUAGUGGA---UGGCGUGGGUGGCGC-AGUGGAAAGUGCUGGGGUAAUGGGA ......((((((.(((.((.(((..(......)......(((((....)))))....))).---)).))).))))))((-(.(....).)))............. ( -31.80) >DroEre_CAF1 983 85 - 1 AAAGUUGCCACUGAUG-CGGCCAAGGAAAAUGCAGAAAAGCU-GCAAACUGGCAUAGCGGCUGG------------------UGGAAAUUGCUGGGGUAGUGGGC .......((((((.(.-((((.........(((((.....))-))).((..((......))..)------------------).......)))).).)))))).. ( -26.90) >consensus AAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCGUAGUGGA___UGGCGUGGGUGGCGC_AGUGGAAAGUGCUGGGGUAAUGGGA ...((((((.....((.((((..................)))).))..(..(((............((((.....))))..........)))..))))))).... (-18.09 = -18.34 + 0.25)


| Location | 11,961,158 – 11,961,260 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.82 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -24.59 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11961158 102 - 22224390 GGGGGUUUUCAUCCACGGUAGCUCUUUUUUUGGUUAAUGAAAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCUUAGUGGA------------------U ..........((((((....((..(((((((((((..(....)...((((....)))))))))))))))..))....(((((((....))))))).)))))------------------) ( -34.40) >DroSec_CAF1 444 102 - 1 GGGAGUUUUCAUCCACGGUAGCUCUUUUUUUGGUUAAUGAAAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCGUAGUGGA------------------U ..........((((((....((..(((((((((((..(....)...((((....)))))))))))))))..))......(((((....)))))...)))))------------------) ( -32.80) >DroSim_CAF1 445 102 - 1 GGGGGUUCUCAUCCACGGUAGCUCUUUUUUUGGUUAAUGAAAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCGUAGUGGA------------------U (((....)))((((((....((..(((((((((((..(....)...((((....)))))))))))))))..))......(((((....)))))...)))))------------------) ( -33.70) >DroEre_CAF1 1006 102 - 1 GGUGGUUUCCAUCUACGGUAGCUCUUUUUUUGGUUAAUGAAAAGUUGCCACUGAUG-CGGCCAAGGAAAAUGCAGAAAAGCU-GCAAACUGGCAUAGCGGCUGG---------------- ..(((((..((((...(((((((.(((.((.....)).))).)))))))...))))-.))))).......(((((.....))-)))..(..((......))..)---------------- ( -27.50) >DroYak_CAF1 1005 111 - 1 GGUGGUUUUCAUCUACGGUAGCUCUUU-UUUGGUUAAUGAAAAGUUGCCACUGAUGGCGACCAAAGAAAAUGCAGAAAAGC--------UGGCGUUGUGGAUGGUUUGGAUGGCGUGGAU .((.(((((((((((((...(((((((-((((...........(((((((....)))))))...........)))))))).--------.)))..)))))))))...)))).))...... ( -31.35) >consensus GGGGGUUUUCAUCCACGGUAGCUCUUUUUUUGGUUAAUGAAAAGUUGCCACUGAUGGCGGCCAAAGAAAAUGCAGAAAAGCCGGCAAACUGGCGUAGUGGA__________________U ...........(((((....((..(((((((((((..(....)...((((....)))))))))))))))..))......(((((....)))))...)))))................... (-24.59 = -25.36 + 0.77)


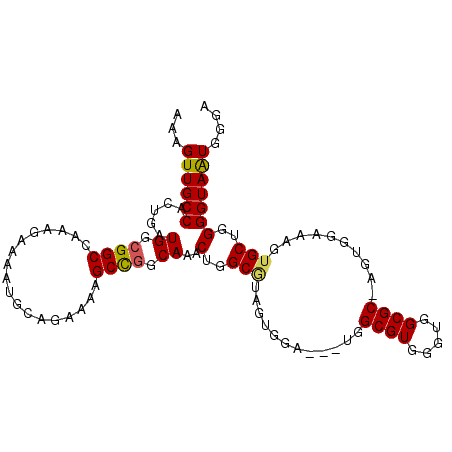
| Location | 11,961,220 – 11,961,336 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11961220 116 + 22224390 UCAUUAACCAAAAAAAGAGCUACCGUGGAUGAAAACCCCCAUCAUACAUGCUAAAAA-AAAAGACACUUGCCAAUGAACACUGGCUGAAAU-UAAAAAAAAAAAUAGUUUUACUUACA ...............(((((((..(((((((........)))).)))..........-...........((((........))))......-............)))))))....... ( -14.50) >DroEre_CAF1 1068 113 + 1 UCAUUAACCAAAAAAAGAGCUACCGUAGAUGGAAACCACCAUCAUGCAUGCUUAAAA--AAAGACACUUGCCAGUGGACACUGGCAAAAAUU-UGGAAAAAA--UUGUUUCACUUUCC .......(((((....((((...(((.(((((......))))))))...))))....--........((((((((....))))))))...))-)))......--.............. ( -28.60) >DroYak_CAF1 1077 113 + 1 UCAUUAACCAAA-AAAGAGCUACCGUAGAUGAAAACCACCAUCAUGUAUGCUCAAAAAAAAAAACACUUGCCAAAGGACACUGGCAAAAAA--UGGUGACAA--UAGUUUUCCUUAUC ............-...((((...(((.((((........)))))))...)))).......(((((..((((((........))))))....--((....)).--..)))))....... ( -22.80) >consensus UCAUUAACCAAAAAAAGAGCUACCGUAGAUGAAAACCACCAUCAUGCAUGCUAAAAA_AAAAGACACUUGCCAAUGGACACUGGCAAAAAU__UGGAAAAAA__UAGUUUUACUUACC ................((((...(((.((((........)))))))...))))..............((((((........))))))............................... (-14.03 = -14.70 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:46 2006