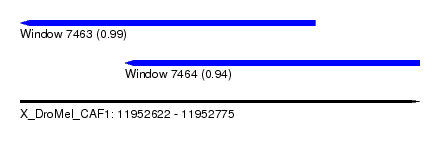
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,952,622 – 11,952,775 |
| Length | 153 |
| Max. P | 0.988352 |

| Location | 11,952,622 – 11,952,735 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.60 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -30.62 |
| Energy contribution | -31.50 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11952622 113 - 22224390 CAGGCCAAAUGGGGGAAUCCCGU---CUCCUUUGUGUGAUUCCCCUUUUUUUUGCCAGCAGCAGCAUUUGGCAU--GCGACCAUCACUAAAUCAGAAUCACAAACGCAUAUCACUCAA ...((((((((((((((((.((.---(......))).))))))))......((((.....))))))))))))((--(((.........................)))))......... ( -31.31) >DroSim_CAF1 20069 114 - 1 CAGGCAAAAUGGGGGAAUCCCGCCGCCGCCUUUGUGUGAUUCCCCUUU--UUUGCCAGCUGCAGCGUUUGGCAU--GCGACCAUCACUAAAUCAGAAUCACAAACGCAUAUCACUCAA ..(((((((.(((((((((.(((..........))).))))))))).)--)))))).......(((((((....--.(................).....)))))))........... ( -36.89) >DroEre_CAF1 20011 111 - 1 CAGGCGAAAUGGGGGAAUCCCGG---CUCUUUUGUGUGAUUCCCCUUU--UUUGCCAACUGCAGCGUUUGGCAU--GCGACCAUCACUAAAUCAGAAUCACAAACGCAUAUCAUUCAA ..(((((((.(((((((((.((.---(......))).))))))))).)--)))))).......(((((((....--.(................).....)))))))........... ( -35.09) >DroYak_CAF1 19378 114 - 1 CAGGCAAAAUGGGGGAAUCCCGU---CUCUUUUGUGUGAUUCCCCUUU-UUUUGCCAGCUGCAGCGUUUGGCAUUGGCGACCAUCACUAAAUCAGAAUCACAAACGCAUAUCACUCAA ..(((((((.(((((((((.((.---(......))).)))))))))..-))))))).......(((((((((....))(.....)...............)))))))........... ( -36.10) >consensus CAGGCAAAAUGGGGGAAUCCCGU___CUCCUUUGUGUGAUUCCCCUUU__UUUGCCAGCUGCAGCGUUUGGCAU__GCGACCAUCACUAAAUCAGAAUCACAAACGCAUAUCACUCAA ..(((((((.(((((((((.((............)).)))))))))...))))))).......(((((((((....))(.....)...............)))))))........... (-30.62 = -31.50 + 0.88)

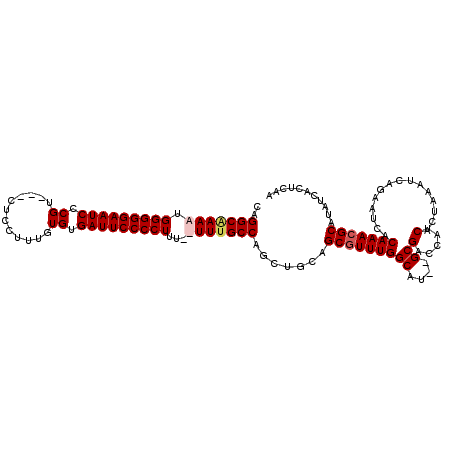
| Location | 11,952,662 – 11,952,775 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -30.13 |
| Energy contribution | -31.32 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11952662 113 - 22224390 AAACAGAGAGUGAAAUCAGAGAAUCAAAACCAUUCUCGCACAGGCCAAAUGGGGGAAUCCCGU---CUCCUUUGUGUGAUUCCCCUUUUUUUUGCCAGCAGCAGCAUUUGGCAU--GC .....(((((((..................)))))))(((...((((((((((((((((.((.---(......))).))))))))......((((.....)))))))))))).)--)) ( -37.27) >DroSim_CAF1 20109 114 - 1 AAACAGAGAGUGAAAUCAGAGAACCAAAACCAUUCUCGCACAGGCAAAAUGGGGGAAUCCCGCCGCCGCCUUUGUGUGAUUCCCCUUU--UUUGCCAGCUGCAGCGUUUGGCAU--GC ((((.(((((((..................)))))))((((.(((((((.(((((((((.(((..........))).))))))))).)--)))))).).)))...)))).....--.. ( -39.97) >DroEre_CAF1 20051 111 - 1 AAACAGAGAGUGAAAUCAGAGAACCAAAACCAUUCUCGCACAGGCGAAAUGGGGGAAUCCCGG---CUCUUUUGUGUGAUUCCCCUUU--UUUGCCAACUGCAGCGUUUGGCAU--GC ((((.(((((((..................)))))))(((..(((((((.(((((((((.((.---(......))).))))))))).)--))))))...)))...)))).....--.. ( -37.87) >DroYak_CAF1 19418 114 - 1 AAACAGAGGCUGAAAUCAGAGAAACAAAACCAUUCCCGCACAGGCAAAAUGGGGGAAUCCCGU---CUCUUUUGUGUGAUUCCCCUUU-UUUUGCCAGCUGCAGCGUUUGGCAUUGGC .........(((....))).....(((..(((..((.((((.(((((((.(((((((((.((.---(......))).)))))))))..-))))))).).))).).)..)))..))).. ( -36.00) >consensus AAACAGAGAGUGAAAUCAGAGAACCAAAACCAUUCUCGCACAGGCAAAAUGGGGGAAUCCCGU___CUCCUUUGUGUGAUUCCCCUUU__UUUGCCAGCUGCAGCGUUUGGCAU__GC .....(((((((..................)))))))(((..(((((((.(((((((((.((............)).)))))))))...)))))))...))).((.....))...... (-30.13 = -31.32 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:37 2006