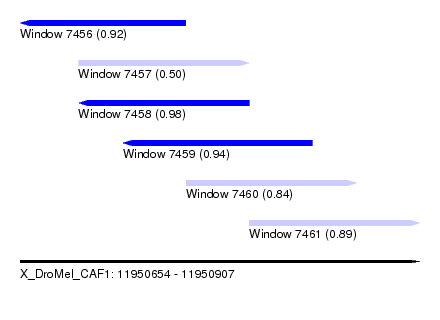
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,950,654 – 11,950,907 |
| Length | 253 |
| Max. P | 0.982691 |

| Location | 11,950,654 – 11,950,759 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.37 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -12.06 |
| Energy contribution | -14.58 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11950654 105 - 22224390 UUCAGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGCCAAUCGAUG-ACGCUGGCUUAAAGCCC-----------UGCCGA--CUGGCUUUAA-AAUGCAAUUUGGAAACAAACAGCGA ....((((..(((((..(((((....)))))...)))))))))....-.(((((((((((((((.-----------......--..))))))))-...))..((((....))))))))). ( -38.50) >DroSec_CAF1 39282 106 - 1 UUCGGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGCCAAUCGAUG-CCGCUGGCUUAAAGC-C-----------UGCCGACACUGGCUUUAA-AAUGCAAUUUGGAAACAAACAGCGA ....((((..(((((..(((((....)))))...)))))))))....-.((((((((((((((-(-----------..........))))))))-...))..((((....))))))))). ( -39.20) >DroEre_CAF1 18083 97 - 1 UUCAGAUUUAGGCCACUUCCGGAUUUCUGGUCCAUGGGCAAUCGAUG-CCGGCGGCUUAAAUU-C-----------UGUCGA--CUGGCUUUAA-AAUGCAAUUUUCAAACAA------- ..((((((((((((.((...((((.....))))...((((.....))-)))).)))))))).)-)-----------))....--...((.....-...)).............------- ( -25.70) >DroYak_CAF1 17785 116 - 1 UUCAGAUUUAGGCCACUUCCGGAAUUCUGGUCCAUGGGCAAUCGAUG-CCGCUGGCUUAAAGC-CGAUGCGAUGCCGGCCGA--CUGGAUUUAAAAAUGCAAUUUUGAAACAAACAGCGA (((((.....((((....((((....))))......((((.(((...-(.(((.......)))-.)...)))))))))))..--)))))(((((((......)))))))........... ( -30.70) >DroAna_CAF1 13195 116 - 1 -ACGGAUUUAGGCCAUUUCCGGAAUGCACGGCGAACGGCAAUCGAUGGCCGGACGCUUAAAGC-CUCCCAGCUGGCGAGUGA-AGUGGCUUUAA-AAUGCAAUUUGGCAACAAACUGCGA -.....((((((((((((((.........((((..((((........))))..))))....((-(........)))..).))-)))))).))))-).((((.((((....)))).)))). ( -40.70) >consensus UUCAGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGGCAAUCGAUG_CCGCUGGCUUAAAGC_C___________UGCCGA__CUGGCUUUAA_AAUGCAAUUUGGAAACAAACAGCGA ......((((((((....((((....)))).....((.((.....)).))...))))))))................((((....))))........(((..((((....))))..))). (-12.06 = -14.58 + 2.52)

| Location | 11,950,691 – 11,950,799 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -14.46 |
| Energy contribution | -17.22 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.38 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11950691 108 + 22224390 GGCA-----------GGGCUUUAAGCCAGCGU-CAUCGAUUGGCCAUGGUCCAGAAUUCCGGAAGUGGCCUAAAUCUGAACAAGCUGGCUUCGUUAAUCGUUUUCGGUUUGCCAGCUCGU ....-----------(((((..((((((((.(-((..((((((((((..(((.(....).))).))))))..)))))))....)))))))).((.(((((....))))).)).))))).. ( -41.30) >DroSec_CAF1 39321 107 + 1 GGCA-----------G-GCUUUAAGCCAGCGG-CAUCGAUUGGCCAUGGUCCAGAAUUCCGGAAGUGGCCUAAAUCCGAACAAGCUGGCUUCGUUAAUCGUUUUCGGUUUGCCAGCUCGU ((((-----------(-(((..((((((((..-..((((((((((((..(((.(....).))).))))))..))).)))....))))))))((.....)).....))))))))....... ( -41.20) >DroEre_CAF1 18113 107 + 1 GACA-----------G-AAUUUAAGCCGCCGG-CAUCGAUUGCCCAUGGACCAGAAAUCCGGAAGUGGCCUAAAUCUGAACAAGCUGGCUUCGUUAAUCGUUUACGGUUUGCCAGCUCGU ..((-----------(-(.((((.(((((.((-((.....))))..((((.......))))...))))).)))))))).((.(((((((.((((.........))))...))))))).)) ( -37.60) >DroYak_CAF1 17823 118 + 1 GGCCGGCAUCGCAUCG-GCUUUAAGCCAGCGG-CAUCGAUUGCCCAUGGACCAGAAUUCCGGAAGUGGCCUAAAUCUGAACAAGCUGGCUUUGUUAAUCGUUUUCGGUUUGCCAGCUCGU (((((.(.((((...(-((.....))).))((-((.....))))..((((.......)))))).)))))).........((.(((((((......(((((....))))).))))))).)) ( -37.60) >DroAna_CAF1 13233 102 + 1 ACUCGCCAGCUGGGAG-GCUUUAAGCGUCCGGCCAUCGAUUGCCGUUCGCCGUGCAUUCCGGAAAUGGCCUAAAUCCGU-----------------AUCGUUUACGGUUUGUCAGCUCAG ....((((.((((((.-((.....(((..((((........))))..)))...)).))))))...)))).((((.((((-----------------(.....)))))))))......... ( -34.90) >consensus GGCA___________G_GCUUUAAGCCAGCGG_CAUCGAUUGCCCAUGGACCAGAAUUCCGGAAGUGGCCUAAAUCUGAACAAGCUGGCUUCGUUAAUCGUUUUCGGUUUGCCAGCUCGU (((...................((((((((.......((((..((((...((.(....).))..))))....)))).......))))))))....(((((....))))).)))....... (-14.46 = -17.22 + 2.76)

| Location | 11,950,691 – 11,950,799 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -19.04 |
| Energy contribution | -22.04 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11950691 108 - 22224390 ACGAGCUGGCAAACCGAAAACGAUUAACGAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGCCAAUCGAUG-ACGCUGGCUUAAAGCCC-----------UGCC ....((.(((..........((.....))(((((((((..((..((((..(((((..(((((....)))))...)))))))))))..-).))))))))...))).-----------.)). ( -38.50) >DroSec_CAF1 39321 107 - 1 ACGAGCUGGCAAACCGAAAACGAUUAACGAAGCCAGCUUGUUCGGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGCCAAUCGAUG-CCGCUGGCUUAAAGC-C-----------UGCC ....((.(((..........((.....))((((((((....(((.(((..(((((..(((((....)))))...)))))))))))..-..))))))))...))-)-----------.)). ( -39.30) >DroEre_CAF1 18113 107 - 1 ACGAGCUGGCAAACCGUAAACGAUUAACGAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAUUUCUGGUCCAUGGGCAAUCGAUG-CCGGCGGCUUAAAUU-C-----------UGUC ((((((((((....(((.........)))..)))))))))).((((((((((((.((...((((.....))))...((((.....))-)))).)))))))).)-)-----------)).. ( -40.80) >DroYak_CAF1 17823 118 - 1 ACGAGCUGGCAAACCGAAAACGAUUAACAAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAAUUCUGGUCCAUGGGCAAUCGAUG-CCGCUGGCUUAAAGC-CGAUGCGAUGCCGGCC ((((((((((.....................)))))))))).....(((((((((...((((....))))......((((.....))-))..)))))))))((-((.........)))). ( -40.20) >DroAna_CAF1 13233 102 - 1 CUGAGCUGACAAACCGUAAACGAU-----------------ACGGAUUUAGGCCAUUUCCGGAAUGCACGGCGAACGGCAAUCGAUGGCCGGACGCUUAAAGC-CUCCCAGCUGGCGAGU ...(((((.....(((((.....)-----------------)))).....(((((((....((.(((.((.....))))).)))))))))(((.((.....))-.))))))))....... ( -34.60) >consensus ACGAGCUGGCAAACCGAAAACGAUUAACGAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGGCAAUCGAUG_CCGCUGGCUUAAAGC_C___________UGCC ((((((((((.....................)))))))))).....((((((((....((((....)))).....((.((.....)).))...))))))))................... (-19.04 = -22.04 + 3.00)

| Location | 11,950,719 – 11,950,839 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -28.69 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11950719 120 - 22224390 AUAACAAUGCAACACCCAAAUGCAUGGAAUUUUUAAAAAUACGAGCUGGCAAACCGAAAACGAUUAACGAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGCCA ......(((((.........)))))...............((((((((((.....................)))))))))).........(((((..(((((....)))))...))))). ( -34.40) >DroSec_CAF1 39348 120 - 1 AUAACAAUGCGACACCCAAAUGCAUGGAAUUUUUAAAAAUACGAGCUGGCAAACCGAAAACGAUUAACGAAGCCAGCUUGUUCGGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGCCA .....(((.(((...(((......))).............((((((((((.....................))))))))))))).)))..(((((..(((((....)))))...))))). ( -36.30) >DroEre_CAF1 18140 120 - 1 AUAACUAUGCAACACCCAAAUGCAUGGAAUUUUUAAAAAUACGAGCUGGCAAACCGUAAACGAUUAACGAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAUUUCUGGUCCAUGGGCA ....(((((((.........))))))).....(((((...((((((((((....(((.........)))..)))))))))).....)))))(((....((((....))))......))). ( -34.50) >DroYak_CAF1 17861 120 - 1 AUAACAAUGCAAAACCCAAAUGCAUGGAAUAUUUAAAAAUACGAGCUGGCAAACCGAAAACGAUUAACAAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAAUUCUGGUCCAUGGGCA ......................((((((............((((((((((.....................))))))))))(((((.((.((......))..)).))))))))))).... ( -29.00) >consensus AUAACAAUGCAACACCCAAAUGCAUGGAAUUUUUAAAAAUACGAGCUGGCAAACCGAAAACGAUUAACGAAGCCAGCUUGUUCAGAUUUAGGCCACUUCCGGAAUUCUGGACCAUGGCCA ....(.(((((.........))))).).............((((((((((.....................)))))))))).........(((((...((((....))))....))))). (-28.69 = -29.00 + 0.31)

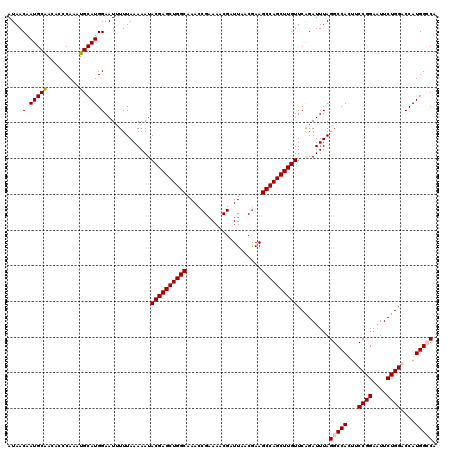
| Location | 11,950,759 – 11,950,867 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.62 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -23.91 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11950759 108 + 22224390 CAAGCUGGCUUCGUUAAUCGUUUUCGGUUUGCCAGCUCGUAUUUUUAAAAAUUCCAUGCAUUUGGGUGUUGCAUUGUUAUCGCCAGAUGU--GUGCGAUGAAA----------AAACAGC ..(((((((......(((((....))))).))))))).((.((((((.....(((((((((((((.((..((...))...))))))))))--))).)))))))----------).))... ( -30.30) >DroSec_CAF1 39388 118 + 1 CAAGCUGGCUUCGUUAAUCGUUUUCGGUUUGCCAGCUCGUAUUUUUAAAAAUUCCAUGCAUUUGGGUGUCGCAUUGUUAUGGCCAGAUGU--GUGCAAUGAAAAAAACACAAAAAACAGC ..(((((((......(((((....))))).))))))).((.(((((....(((.(((((((((((.(((.((...)).))).))))))))--))).))).))))).))............ ( -32.30) >DroEre_CAF1 18180 107 + 1 CAAGCUGGCUUCGUUAAUCGUUUACGGUUUGCCAGCUCGUAUUUUUAAAAAUUCCAUGCAUUUGGGUGUUGCAUAGUUAUGGCCAGAUGU--GUGCAAUGAAA----------A-ACAGC ..(((((((.((((.........))))...)))))))....(((((....(((.(((((((((((.(((.((...)).))).))))))))--))).))).)))----------)-).... ( -30.50) >DroYak_CAF1 17901 110 + 1 CAAGCUGGCUUUGUUAAUCGUUUUCGGUUUGCCAGCUCGUAUUUUUAAAUAUUCCAUGCAUUUGGGUUUUGCAUUGUUAUGGCCCGAUGUGUGUGCAAUGAAA----------AUAUAGC ..(((((((......(((((....))))).))))))).((((((((....(((.(((((((((((((..((......))..)))))).))))))).)))))))----------))))... ( -31.90) >consensus CAAGCUGGCUUCGUUAAUCGUUUUCGGUUUGCCAGCUCGUAUUUUUAAAAAUUCCAUGCAUUUGGGUGUUGCAUUGUUAUGGCCAGAUGU__GUGCAAUGAAA__________AAACAGC ..(((((((......(((((....))))).))))))).............(((.(((((((((((.(((.((...)).))).))))))))..))).)))..................... (-23.91 = -24.47 + 0.56)

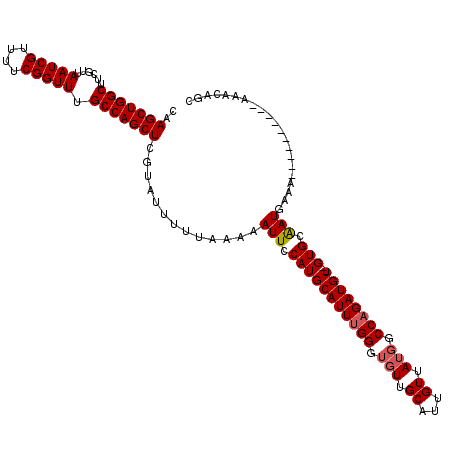

| Location | 11,950,799 – 11,950,907 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11950799 108 + 22224390 AUUUUUAAAAAUUCCAUGCAUUUGGGUGUUGCAUUGUUAUCGCCAGAUGU--GUGCGAUGAAA----------AAACAGCAAUGCAUACAAUUUUUAAACAAUGCUAACACCAUUGCAAA ................((((....(((((((((((((((((....)))((--(((((.((...----------......)).)))))))........)))))))).))))))..)))).. ( -32.90) >DroSec_CAF1 39428 118 + 1 AUUUUUAAAAAUUCCAUGCAUUUGGGUGUCGCAUUGUUAUGGCCAGAUGU--GUGCAAUGAAAAAAACACAAAAAACAGCAAUGCAUACAAUUUUUAAACAUUGAUAACACCAUUGCAAA ................((((....(((((.(((((((..((......(((--((............))))).....)))))))))...((((........))))...)))))..)))).. ( -24.60) >DroEre_CAF1 18220 107 + 1 AUUUUUAAAAAUUCCAUGCAUUUGGGUGUUGCAUAGUUAUGGCCAGAUGU--GUGCAAUGAAA----------A-ACAGCAAUGCAUACAAUUUUUAGGCACUUCUAACACCUUUGUGAA ..................(((..((((((((...(((....(((((((((--(((((.((...----------.-....)).))))))))...))).))))))..))))))))..))).. ( -27.80) >DroYak_CAF1 17941 110 + 1 AUUUUUAAAUAUUCCAUGCAUUUGGGUUUUGCAUUGUUAUGGCCCGAUGUGUGUGCAAUGAAA----------AUAUAGCAAUGCAUACAAUUUUAAAACAUUGCAAACACCUUUGUAAA ................((((...((((((((((.((((.(((...(((.((((((((.((...----------......)).)))))))))))))).)))).)))))).)))).)))).. ( -26.30) >consensus AUUUUUAAAAAUUCCAUGCAUUUGGGUGUUGCAUUGUUAUGGCCAGAUGU__GUGCAAUGAAA__________AAACAGCAAUGCAUACAAUUUUUAAACAUUGCUAACACCAUUGCAAA ................((((...((((((((((.((((........((((...(((......................)))..))))..........)))).))).))))))).)))).. (-16.86 = -17.79 + 0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:34 2006