| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,948,658 – 11,948,785 |
| Length | 127 |
| Max. P | 0.895792 |

| Location | 11,948,658 – 11,948,758 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
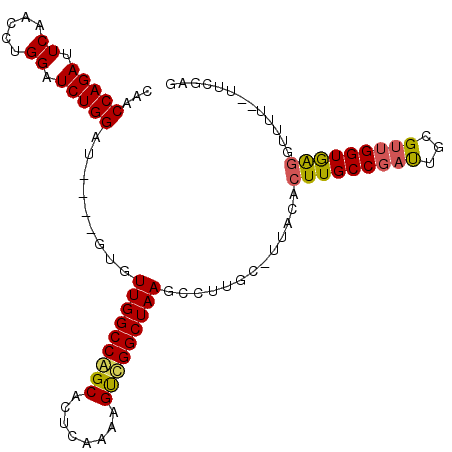
>X_DroMel_CAF1 11948658 100 + 22224390 CGUACCCGCCCACUGACCCAC--UCAAUCACAACCAGAUUCAACCUG---GAUCUGGAU----GUGUUGGCCAGCGCUCAAAAGUCGGCUAAGCCUUGC-UUACGCUUGC ((((...((............--.....((((.((((((((.....)---))))))).)----)))((((((.((........)).)))))).....))-.))))..... ( -27.70) >DroSec_CAF1 31788 101 + 1 CGUACCCGCCCACUGACCCAC--UCAAUCACAACCAGAUUCAACCUG---GUUCUGGAU----GUGUUGGCCGGCACUCAAAAGUCGGCUAAGCCUUGCCUUACACUUGC .(((...((............--.....((((.(((((..(.....)---..))))).)----)))(((((((((........))))))))).....))..)))...... ( -27.60) >DroEre_CAF1 16181 88 + 1 CGUUCCCGCCCACUGACCCAC--UCAAUCACAACCAGACUCAACCUGGGGGAACUGGUC----GUGUUGGCCAGCACUCAAAAGUUGGCUAAAU---------------- .((((((......(((.....--))).......((((.......)))))))))).....----...(((((((((........)))))))))..---------------- ( -27.40) >DroYak_CAF1 15888 106 + 1 CGUUCCCGCCCACUGACCCACUCUCAAUCACAACCAGACUCCGCCUG---GAUCUGGUUAUGUGUGUUGGCCAGCACUCAAAAGCUGGCUAAGCCCUGC-UUACACUUGC ........................((((((((((((((.(((....)---))))))))..)))).))))((((((........))))))(((((...))-)))....... ( -30.20) >consensus CGUACCCGCCCACUGACCCAC__UCAAUCACAACCAGACUCAACCUG___GAUCUGGAU____GUGUUGGCCAGCACUCAAAAGUCGGCUAAGCCUUGC_UUACACUUGC ..........((((((.......))).......(((((..............)))))......)))(((((((((........))))))))).................. (-17.65 = -17.71 + 0.06)


| Location | 11,948,686 – 11,948,785 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -25.23 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11948686 99 + 22224390 CAACCAGAUUCAACCUGGAUCUGGAU----GUGUUGGCCAGCGCUCAAAAGUCGGCUAAGCCUUGC-UUACGCUUGCCGAUUGCGUUGGUAAGGUUUU--UUCGAA ...((((((((.....))))))))..----(((((....))))).......((((..(((((((((-(.((((.........)))).)))))))))).--.)))). ( -38.40) >DroSec_CAF1 31816 100 + 1 CAACCAGAUUCAACCUGGUUCUGGAU----GUGUUGGCCGGCACUCAAAAGUCGGCUAAGCCUUGCCUUACACUUGCCGAUUGCGUUGGUGAAGUUCU--UUCAAG ...(((((..(.....)..)))))..----...(((((((((........)))))))))..((((......((((((((((...)))))).))))...--..)))) ( -28.80) >DroYak_CAF1 15918 105 + 1 CAACCAGACUCCGCCUGGAUCUGGUUAUGUGUGUUGGCCAGCACUCAAAAGCUGGCUAAGCCCUGC-UUACACUUGCCGUCUCCGUAGGUGGGUUUUUUGUUCGAG .....((((.(((((((((..((((...((((((((((((((........)))))))))((...))-.)))))..))))..)))..)))))))))).......... ( -38.90) >consensus CAACCAGAUUCAACCUGGAUCUGGAU____GUGUUGGCCAGCACUCAAAAGUCGGCUAAGCCUUGC_UUACACUUGCCGAUUGCGUUGGUGAGGUUUU__UUCGAG ...(((((.((.....)).))))).........(((((((((........))))))))).............(((((((((...)))))))))............. (-25.23 = -25.47 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:29 2006