| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,944,496 – 11,944,656 |
| Length | 160 |
| Max. P | 0.997231 |

| Location | 11,944,496 – 11,944,616 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11944496 120 + 22224390 UACCAUGCCAUCCUUUGGCACUUCAAGUCGAUUUACAGCUACAAACGAAUGUCUCGAGGACAUUUCCAUGCCUUCAUAUCGCGAUAAUACCGAAUCGCUUAGGCAUAGAUGUCCUACACC .....(((((.....))))).........................(((.....)))(((((((((..((((((.......(((((........)))))..)))))))))))))))..... ( -31.20) >DroSec_CAF1 27506 120 + 1 UACCAUGCCAUCCUAUGGCACUUCAAGUCGAUUUACAGCUACAAAUGAAUGUCUCGAGGACAUUUCCAUGCCUUCAUAUCGCGAUAAUACCGGAUCGCUUAGGCAAAGAUGUCCUACACC .....((((((...)))))).......((((...(((............))).))))((((((((...(((((.......(((((........)))))..))))).))))))))...... ( -29.00) >DroEre_CAF1 12427 120 + 1 UACCAUGCCAUCCUUUGGCACUUCUACCUUUGGCACGGCUACCAAUGAAUGUCUCGAGGACAUUUCAAUGCCUUCAUAUCGGGAUCACACCGGAUCGCUUAGGCAUAGAUGUCCAACACC ..((.(((((......((........))..))))).))...................((((((((..((((((....(((.((......)).))).....))))))))))))))...... ( -32.60) >DroYak_CAF1 12001 120 + 1 UGCCAUGGCAUCCUUUGGUACUUUAAGUCGACAUACAGCUACAAAUGAAUGUCUCGAGGACAUUUCAAUGCCUUCAUAUCGGGAUAAUACCGGAUCGCUGAGGCAUAGAUGUCCAACACC (((((.((...))..))))).........(((((.((........)).)))))....((((((((..(((((((...(((.((......)).)))....)))))))))))))))...... ( -34.50) >consensus UACCAUGCCAUCCUUUGGCACUUCAAGUCGAUUUACAGCUACAAAUGAAUGUCUCGAGGACAUUUCAAUGCCUUCAUAUCGCGAUAAUACCGGAUCGCUUAGGCAUAGAUGUCCAACACC .....(((((.....))))).....................................((((((((..((((((.....((((.......)))).......))))))))))))))...... (-23.00 = -23.62 + 0.62)


| Location | 11,944,536 – 11,944,656 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -19.96 |
| Energy contribution | -21.27 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11944536 120 + 22224390 ACAAACGAAUGUCUCGAGGACAUUUCCAUGCCUUCAUAUCGCGAUAAUACCGAAUCGCUUAGGCAUAGAUGUCCUACACCAAUUCAGAGCUCUAGACAAGAAUGUCUGGAUGAUAUGACU ......((((......(((((((((..((((((.......(((((........)))))..)))))))))))))))......)))).....((((((((....)))))))).......... ( -36.30) >DroSec_CAF1 27546 91 + 1 ACAAAUGAAUGUCUCGAGGACAUUUCCAUGCCUUCAUAUCGCGAUAAUACCGGAUCGCUUAGGCAAAGAUGUCCUACACCAAUUCAGAGCU----------------------------- .....(((((......(((((((((...(((((.......(((((........)))))..))))).)))))))))......))))).....----------------------------- ( -26.50) >DroEre_CAF1 12467 91 + 1 ACCAAUGAAUGUCUCGAGGACAUUUCAAUGCCUUCAUAUCGGGAUCACACCGGAUCGCUUAGGCAUAGAUGUCCAACACCAAUGCAGAGCU----------------------------- ..........(.(((..((((((((..((((((....(((.((......)).))).....))))))))))))))..((....))..)))).----------------------------- ( -23.50) >DroYak_CAF1 12041 91 + 1 ACAAAUGAAUGUCUCGAGGACAUUUCAAUGCCUUCAUAUCGGGAUAAUACCGGAUCGCUGAGGCAUAGAUGUCCAACACCAAUUCAGAGCA----------------------------- .....(((((.......((((((((..(((((((...(((.((......)).)))....))))))))))))))).......))))).....----------------------------- ( -26.94) >consensus ACAAAUGAAUGUCUCGAGGACAUUUCAAUGCCUUCAUAUCGCGAUAAUACCGGAUCGCUUAGGCAUAGAUGUCCAACACCAAUUCAGAGCU_____________________________ ......((((......(((((((((..((((((.....((((.......)))).......)))))))))))))))......))))................................... (-19.96 = -21.27 + 1.31)

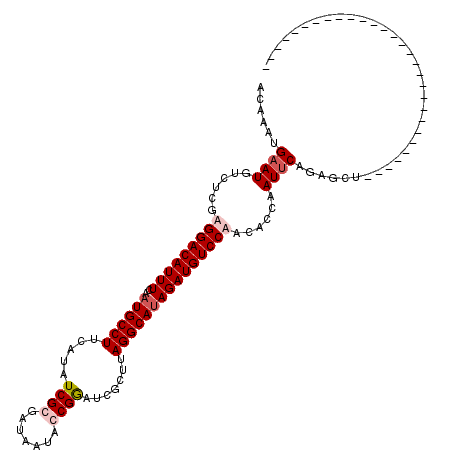

| Location | 11,944,536 – 11,944,656 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -21.02 |
| Energy contribution | -22.40 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11944536 120 - 22224390 AGUCAUAUCAUCCAGACAUUCUUGUCUAGAGCUCUGAAUUGGUGUAGGACAUCUAUGCCUAAGCGAUUCGGUAUUAUCGCGAUAUGAAGGCAUGGAAAUGUCCUCGAGACAUUCGUUUGU .(((((((((..((((..((((.....)))).))))...))))))(((((((((((((((..(((((........))))).......)))))))))..))))))...))).......... ( -38.20) >DroSec_CAF1 27546 91 - 1 -----------------------------AGCUCUGAAUUGGUGUAGGACAUCUUUGCCUAAGCGAUCCGGUAUUAUCGCGAUAUGAAGGCAUGGAAAUGUCCUCGAGACAUUCAUUUGU -----------------------------.....(((((.(.((.(((((((((.(((((..(((((........))))).......))))).)))..))))))))...))))))..... ( -25.00) >DroEre_CAF1 12467 91 - 1 -----------------------------AGCUCUGCAUUGGUGUUGGACAUCUAUGCCUAAGCGAUCCGGUGUGAUCCCGAUAUGAAGGCAUUGAAAUGUCCUCGAGACAUUCAUUGGU -----------------------------.......((.((((((((((((((.((((((.....(((.((......)).)))....)))))).))..)))))....)))).))).)).. ( -23.90) >DroYak_CAF1 12041 91 - 1 -----------------------------UGCUCUGAAUUGGUGUUGGACAUCUAUGCCUCAGCGAUCCGGUAUUAUCCCGAUAUGAAGGCAUUGAAAUGUCCUCGAGACAUUCAUUUGU -----------------------------......((((..((((((((((((.((((((((...(((.((......)).))).)).)))))).))..)))))....)))))..)))).. ( -26.40) >consensus _____________________________AGCUCUGAAUUGGUGUAGGACAUCUAUGCCUAAGCGAUCCGGUAUUAUCCCGAUAUGAAGGCAUGGAAAUGUCCUCGAGACAUUCAUUUGU ...................................((((..((((.((((((((((((((.....(((.((......)).)))....)))))))))..))))).....))))..)))).. (-21.02 = -22.40 + 1.38)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:27 2006