
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,940,475 – 11,940,608 |
| Length | 133 |
| Max. P | 0.950657 |

| Location | 11,940,475 – 11,940,571 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -30.49 |
| Energy contribution | -30.35 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
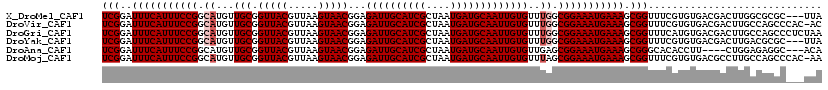
>X_DroMel_CAF1 11940475 96 + 22224390 GG------------------------AGCAGCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGA .(------------------------(((((((((.((((..(((.......))))))).))))).(((((.....)))))...((((((((((....)))))))))).)))))...... ( -34.60) >DroVir_CAF1 28913 96 + 1 GG------------------------CGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGA ..------------------------..(((((((.((((..(((.......)))))))....)).(((((.....)))))...((((((((((....)))))))))).....))))).. ( -33.50) >DroGri_CAF1 4755 96 + 1 GG------------------------CGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGA ..------------------------..(((((((.((((..(((.......)))))))....)).(((((.....)))))...((((((((((....)))))))))).....))))).. ( -33.50) >DroYak_CAF1 7872 96 + 1 GG------------------------AGCAGCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGA .(------------------------(((((((((.((((..(((.......))))))).))))).(((((.....)))))...((((((((((....)))))))))).)))))...... ( -34.60) >DroAna_CAF1 3198 120 + 1 GGCGUGACAGCGAUGGCGACGAGGACGGCGACGGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUGAGCGGA ((((((((.((((((((...((((..(((....)))..))))(((.......))).)).)))))).))))))))...(((((..((((((((((....)))))))))).)))))...... ( -44.40) >DroMoj_CAF1 3826 96 + 1 GG------------------------CGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUAGCGGA ((------------------------(.(((.(((.((((..(((.......))))))).))))))))).(((((((((.....((((((((((....)))))))))).))))))))).. ( -32.60) >consensus GG________________________CGCGCCAGCUUGCCUCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGA ...........................((..((((.((((..(((.......))))))).))))(.(((((.....))))).).((((((((((....)))))))))).......))... (-30.49 = -30.35 + -0.14)



| Location | 11,940,475 – 11,940,571 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -22.95 |
| Energy contribution | -22.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11940475 96 - 22224390 UCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCUGCU------------------------CC ...(((.......((((((((......)))))))).((((((((.....))))).((.....)).)))............))).(((....)))------------------------.. ( -24.60) >DroVir_CAF1 28913 96 - 1 UCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGGCGCG------------------------CC ..(((((......((((((((......))))))))....(((((.....))))).((....(((((((.......)))..)))).)))))))..------------------------.. ( -26.90) >DroGri_CAF1 4755 96 - 1 UCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGGCGCG------------------------CC ..(((((......((((((((......))))))))....(((((.....))))).((....(((((((.......)))..)))).)))))))..------------------------.. ( -26.90) >DroYak_CAF1 7872 96 - 1 UCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGCUGCU------------------------CC ...(((.......((((((((......)))))))).((((((((.....))))).((.....)).)))............))).(((....)))------------------------.. ( -24.60) >DroAna_CAF1 3198 120 - 1 UCCGCUCAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCCGUCGCCGUCCUCGUCGCCAUCGCUGUCACGCC ................((.....(((((((((....((((((((.....))))).((.....)).))).........(((((...((....))...)))))..).))))))))....)). ( -28.30) >DroMoj_CAF1 3826 96 - 1 UCCGCUAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGGCGCG------------------------CC ..(((........((((((((......))))))))....(((((.....))))).((..(.(((((((.......)))..)))).)...)))))------------------------.. ( -24.80) >consensus UCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGAGGCAAGCUGGCGCG________________________CC ...(((.......((((((((......)))))))).((((((((.....))))).((.....)).)))............)))..((....))........................... (-22.95 = -22.73 + -0.22)



| Location | 11,940,491 – 11,940,608 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.11 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -34.33 |
| Energy contribution | -34.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11940491 117 + 22224390 UCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUUUCGUGUGACGACUUGGCGCGC---UUA (((..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).))))))))))).)))....(((((.(......))))))---... ( -41.70) >DroVir_CAF1 28929 119 + 1 UCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUUUCGUGUGACGACUUGCCAGCCCAC-AC ..((.(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))(((((.(((.....)))...))).))))..-.. ( -43.40) >DroGri_CAF1 4771 120 + 1 UCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUUUCAUGUGACGACUUGCCAGCCCUCUAA ..((.(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))))(((((.((.......))...))).))))..... ( -39.80) >DroYak_CAF1 7888 117 + 1 UCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUUUCGUGUGACGACUUGACGCGC---UUA .......(((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).)))))))))(((((((((((.....)))...))).)))---)). ( -42.10) >DroAna_CAF1 3238 113 + 1 UCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUGAGCGGAAAUGAAAGCGGGCACACCUU----CUGGAGAGGC---ACA (((..(((((((((((((((....(.(((((.....))))).).((((((((((....))))))))))))))....))))))))))).)))((.(.((..----..)).)..))---... ( -37.60) >DroMoj_CAF1 3842 119 + 1 UCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUAGCGGAAAUGAAAGCGGUUUCGUGUGACGCCUUGCCAGCCCAC-AA ..((.(((((((((((((((....(.(((((.....))))).).((((((((((....))))))))))))))....)))))))))))(((((...(((...)))...))).))))..-.. ( -39.40) >consensus UCGGAUUUCAUUUCCGGCAUGUUGCGGUUACGUUAAGUAACGGAGAUUGCAUCGCUAAUGAUGCAAUUGUGUUUGGCGGAAAUGAAAGCGGUUUCGUGUGACGACUUGCCAGCC___UAA (((..(((((((((((.((...(((.(((((.....)))))...((((((((((....)))))))))))))..)).))))))))))).)))............................. (-34.33 = -34.83 + 0.50)


| Location | 11,940,491 – 11,940,608 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.11 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -27.81 |
| Energy contribution | -27.75 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
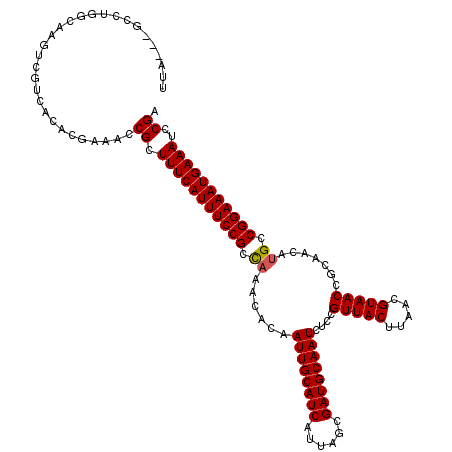
>X_DroMel_CAF1 11940491 117 - 22224390 UAA---GCGCGCCAAGUCGUCACACGAAACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGA ...---..(((.....(((.....)))...)))(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..... ( -33.30) >DroVir_CAF1 28929 119 - 1 GU-GUGGGCUGGCAAGUCGUCACACGAAACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGA ((-(((((((....))))..))))).....((.(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..)). ( -38.10) >DroGri_CAF1 4771 120 - 1 UUAGAGGGCUGGCAAGUCGUCACAUGAAACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGA ...((.((((....)))).)).........((.(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..)). ( -35.90) >DroYak_CAF1 7888 117 - 1 UAA---GCGCGUCAAGUCGUCACACGAAACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGA ..(---(((.((....(((.....))).))))))((((((((((.((......((((((((......))))))))....(((((.....))))).......)).))))))))))...... ( -34.20) >DroAna_CAF1 3238 113 - 1 UGU---GCCUCUCCAG----AAGGUGUGCCCGCUUUCAUUUCCGCUCAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGA .(.---.((((....)----.)))..)...((.(((((((((((.........((((((((......))))))))....(((((.....))))).((.....)))))))))))))..)). ( -31.40) >DroMoj_CAF1 3842 119 - 1 UU-GUGGGCUGGCAAGGCGUCACACGAAACCGCUUUCAUUUCCGCUAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGA .(-(((((((.....))).)))))......((.(((((((((((.........((((((((......))))))))....(((((.....))))).((.....)))))))))))))..)). ( -34.50) >consensus UUA___GCCUGGCAAGUCGUCACACGAAACCGCUUUCAUUUCCGCCAAACACAAUUGCAUCAUUAGCGAUGCAAUCUCCGUUACUUAACGUAACCGCAACAUGCCGGAAAUGAAAUCCGA ..............................((.(((((((((((.((......((((((((......))))))))....(((((.....))))).......)).)))))))))))..)). (-27.81 = -27.75 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:24 2006