
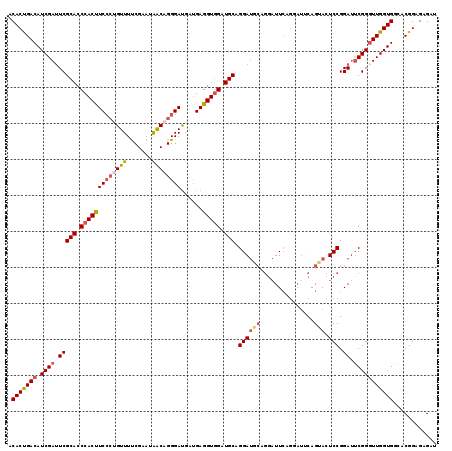
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,935,487 – 11,935,638 |
| Length | 151 |
| Max. P | 0.837622 |

| Location | 11,935,487 – 11,935,599 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -42.86 |
| Consensus MFE | -31.56 |
| Energy contribution | -32.52 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11935487 112 - 22224390 ACACUGACAUCGAUUCGCACCCACUUCCCUGUUUUCGAAUAACAGGGAUGAUGAGGUGGAUGCAGGA-------UUCAGGAUUCAGUACUCCGGAUUCGGGUUGGUGGCACGGAGAGAU ...(((.((((((((((((.(((((((((((((.......))))))))......))))).))).(((-------(((.(((........))))))))))))))))))...)))...... ( -41.50) >DroSec_CAF1 18658 118 - 1 ACACUGACAUCGAUUCGCACCCACUUCCCUGUUUUCGAAUAACAGGGAUGAUGAGGUGGAUGCAGGAUGCAGGAUUCAGUAUUCAGUACUCCGGAUUCGGGUUGGUGG-ACGGAGAGAU .(((..((.((((.(((((.(((((((((((((.......))))))))......))))).))).((((((.((((.....)))).))).))).)).))))))..))).-.......... ( -43.00) >DroSim_CAF1 9349 119 - 1 ACACUGACAUCGAUUCGCACCCACUUCCCUGUUUUCGAAUAACAGGGAUGAUGAGGUGGAUGCAGGAUGCAGGAUUCAGUAUUCAGUACUCCGGAUUCGGGUUGGUGGCACGGAGAGAU .(((..((.((((.(((((.(((((((((((((.......))))))))......))))).))).((((((.((((.....)))).))).))).)).))))))..)))............ ( -43.00) >DroEre_CAF1 3384 119 - 1 ACACCGACAUCGAAUCGCACCCACUUCCAAGUUUCCGAAGGGCAGGGACGACGAGGUGGGUGCAGGAUGCAGGAUUCGGGAUACAGUACUCCGGAACCGGGUUGGUGGAACGGAGAGCU .(((((((.((((((((((((((((((...((((((....))...))))...))))))))))).(....)..)))))))...........(((....))))))))))............ ( -48.80) >DroYak_CAF1 2876 112 - 1 ACACUGACAUCGAUUCGCACCCACCUCGCAGUUUUCGAAGGGCAGGGAUGAUGAGGUUGGUGCAGGAUUCAGGAUU-------CAGAACUCCGGAUUCGGCUUGGUGGCACGGAGAGAU ...((((.(((((((((((((.((((((..((((..(.....)..))))..)))))).))))).)))))...))))-------)))..(((((......(((....))).))))).... ( -38.00) >consensus ACACUGACAUCGAUUCGCACCCACUUCCCUGUUUUCGAAUAACAGGGAUGAUGAGGUGGAUGCAGGAUGCAGGAUUCAGGAUUCAGUACUCCGGAUUCGGGUUGGUGGCACGGAGAGAU .(((((((.((((.(((((.(((((((((((((.......))))))))......))))).))).((((((...............))).))).)).)))))))))))............ (-31.56 = -32.52 + 0.96)


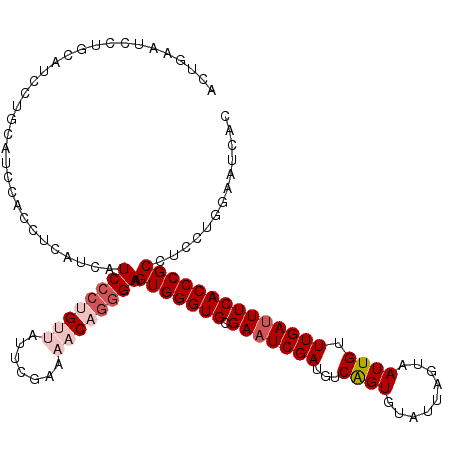
| Location | 11,935,526 – 11,935,638 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -23.14 |
| Energy contribution | -24.78 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11935526 112 + 22224390 CCUGAA-------UCCUGCAUCCACCUCAUCAUCCCUGUUAUUCGAAAACAGGGAAGUGGGUGCGAAUCGAUGUCAGUGUAUUAGUAAUUGUUUGAUUUCACCCGCCUCCUGGUAACAC ......-------..........(((......((((((((.......)))))))).(((((((.(((((((...((((.........)))).)))))))))))))).....)))..... ( -33.10) >DroSec_CAF1 18696 119 + 1 ACUGAAUCCUGCAUCCUGCAUCCACCUCAUCAUCCCUGUUAUUCGAAAACAGGGAAGUGGGUGCGAAUCGAUGUCAGUGUAUUAGUAAUUGUUUGAUUUCACCCGCCUCCUGGAAUCAC ((((((..(((((((.(((((((((.......((((((((.......)))))))).)))))))))....)))).)))..).))))).......((((((((.........)))))))). ( -37.50) >DroSim_CAF1 9388 119 + 1 ACUGAAUCCUGCAUCCUGCAUCCACCUCAUCAUCCCUGUUAUUCGAAAACAGGGAAGUGGGUGCGAAUCGAUGUCAGUGUAUUAGUAAUUGUUUGAUUUCACCCGCCUCCUGGAAUCAC ((((((..(((((((.(((((((((.......((((((((.......)))))))).)))))))))....)))).)))..).))))).......((((((((.........)))))))). ( -37.50) >DroEre_CAF1 3423 110 + 1 CCCGAAUCCUGCAUCCUGCACCCACCUCGUCGUCCCUGCCCUUCGGAAACUUGGAAGUGGGUGCGAUUCGAUGUCGGUGUAUUAGUAAUUGUUUGAUUUCACCCGCCUCC--------- ..((((((.........((((((((.((...((..(((.....)))..))...)).))))))))))))))..((.((((.(((((.......)))))..)))).))....--------- ( -29.90) >DroYak_CAF1 2912 115 + 1 ----AAUCCUGAAUCCUGCACCAACCUCAUCAUCCCUGCCCUUCGAAAACUGCGAGGUGGGUGCGAAUCGAUGUCAGUGUAUUAGUAAUUGUUUGAUUUCACCCGCCUCCGGGAGUCAC ----.....(((.(((((...................((............))((((((((((.(((((((...((((.........)))).)))))))))))))))))))))).))). ( -34.30) >consensus ACUGAAUCCUGCAUCCUGCAUCCACCUCAUCAUCCCUGUUAUUCGAAAACAGGGAAGUGGGUGCGAAUCGAUGUCAGUGUAUUAGUAAUUGUUUGAUUUCACCCGCCUCCUGGAAUCAC ................................((((((((.......)))))))).(((((((.(((((((...((((.........)))).))))))))))))))............. (-23.14 = -24.78 + 1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:20 2006