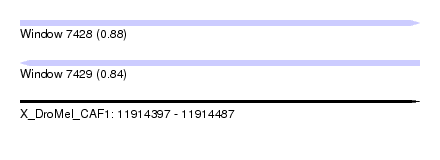
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,914,397 – 11,914,487 |
| Length | 90 |
| Max. P | 0.876714 |

| Location | 11,914,397 – 11,914,487 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11914397 90 + 22224390 GGAACUGAAUCGAAAAGGAUUCAGGAUGGGCGUUGG--------------------------CAAUGGCAAUGGUCGCCUAAUAAGCCAACACAGCAGAGCGUGACAAAACAAACA ((..(((((((......)))))))..((((((...(--------------------------(....))......)))))).....))..(((.((...)))))............ ( -22.80) >DroSec_CAF1 24239 90 + 1 GGAACUGAAUCGAAAAGGAUUCAGGAUGGGCGAUGG--------------------------CAAUGGCAAUGGUCGCCUAAUAAGCCAACACACCAGAGCGUGACAAAACAAACA ((..(((((((......)))))))..((((((((.(--------------------------(....))....)))))))).....))..(((........)))............ ( -25.40) >DroSim_CAF1 22217 90 + 1 GGAACUGAAUCGAAAAGGAUUCAGGAUGGGCGAUGG--------------------------CAAUGGCAAUGGUCGCCUAAUAAGCCAACACACCAGAGCGUGACAAAACAAACA ((..(((((((......)))))))..((((((((.(--------------------------(....))....)))))))).....))..(((........)))............ ( -25.40) >DroEre_CAF1 22514 116 + 1 GGUACUGAAUCGAAAGGGAUUCAGGAUGGGCGAUGGGCGAUGGGCGAUGGUGAGGUUGAUGAUGAUGGCAAUGGUCGCCUAAUAAGCCAACACACCAGAGCGUGACAAAACAAACA (((.(((((((......)))))))....(((..((((((((..((.((.((.(......).)).)).))....))))))))....))).....))).....((......))..... ( -34.90) >DroYak_CAF1 25492 96 + 1 GGUACUAAAUCGAAAGGGAUUUAGGAUGGGCGAUGGA--------------------GAUGGUGAUGGCAAUGUUCGCCUAAUAAGCCAACACACCAGAGCGUGACAAAACAAACA ....(((((((......))))))).....((..(((.--------------------(.((((...(((.......)))......))))...).)))..))((......))..... ( -21.20) >consensus GGAACUGAAUCGAAAAGGAUUCAGGAUGGGCGAUGG__________________________CAAUGGCAAUGGUCGCCUAAUAAGCCAACACACCAGAGCGUGACAAAACAAACA ((..(((((((......)))))))..((((((((.......................................)))))))).....))..(((........)))............ (-19.59 = -19.67 + 0.08)

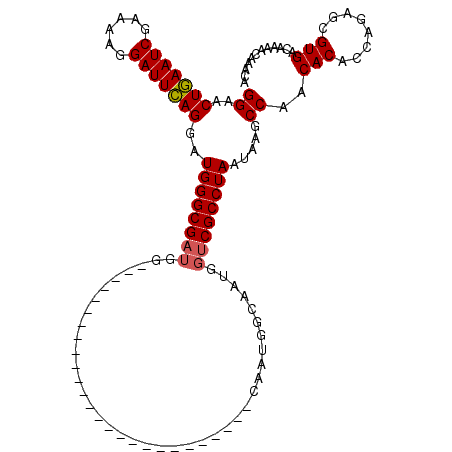

| Location | 11,914,397 – 11,914,487 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.24 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11914397 90 - 22224390 UGUUUGUUUUGUCACGCUCUGCUGUGUUGGCUUAUUAGGCGACCAUUGCCAUUG--------------------------CCAACGCCCAUCCUGAAUCCUUUUCGAUUCAGUUCC .((.((......)).))......((((((((......(((((...)))))...)--------------------------))))))).....(((((((......))))))).... ( -24.20) >DroSec_CAF1 24239 90 - 1 UGUUUGUUUUGUCACGCUCUGGUGUGUUGGCUUAUUAGGCGACCAUUGCCAUUG--------------------------CCAUCGCCCAUCCUGAAUCCUUUUCGAUUCAGUUCC .((.((......)).))..(((.(((.((((......(((((...)))))...)--------------------------))).))))))..(((((((......))))))).... ( -24.40) >DroSim_CAF1 22217 90 - 1 UGUUUGUUUUGUCACGCUCUGGUGUGUUGGCUUAUUAGGCGACCAUUGCCAUUG--------------------------CCAUCGCCCAUCCUGAAUCCUUUUCGAUUCAGUUCC .((.((......)).))..(((.(((.((((......(((((...)))))...)--------------------------))).))))))..(((((((......))))))).... ( -24.40) >DroEre_CAF1 22514 116 - 1 UGUUUGUUUUGUCACGCUCUGGUGUGUUGGCUUAUUAGGCGACCAUUGCCAUCAUCAUCAACCUCACCAUCGCCCAUCGCCCAUCGCCCAUCCUGAAUCCCUUUCGAUUCAGUACC ...............((..(((((.(((((.......(((((...))))).......)))))..)))))..))...................(((((((......))))))).... ( -24.94) >DroYak_CAF1 25492 96 - 1 UGUUUGUUUUGUCACGCUCUGGUGUGUUGGCUUAUUAGGCGAACAUUGCCAUCACCAUC--------------------UCCAUCGCCCAUCCUAAAUCCCUUUCGAUUUAGUACC (((((((((.(((((((......))).)))).....)))))))))..............--------------------.............(((((((......))))))).... ( -18.40) >consensus UGUUUGUUUUGUCACGCUCUGGUGUGUUGGCUUAUUAGGCGACCAUUGCCAUUG__________________________CCAUCGCCCAUCCUGAAUCCUUUUCGAUUCAGUUCC ..........(((((((......))).))))......(((((...)))))..........................................(((((((......))))))).... (-18.56 = -18.24 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:06 2006