
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,912,577 – 11,912,732 |
| Length | 155 |
| Max. P | 0.958391 |

| Location | 11,912,577 – 11,912,692 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.66 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
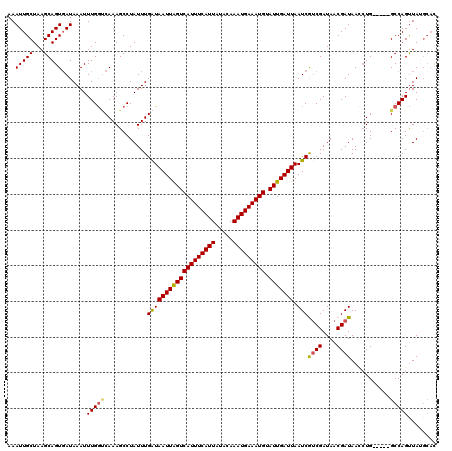
>X_DroMel_CAF1 11912577 115 + 22224390 AAAUUGCUAAGCAGUUAUAAAUUUGAUCAAAGCCUAUUUGAUAAUUGGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCGGCGAUAACGAUAACCUG-----GCCAGUUAUGCAU ..........(((.............(((((.....)))))(((((((((((((((((.....))))))))..........((((.......)))).....)-----))))))))))).. ( -24.40) >DroSec_CAF1 22434 115 + 1 AAAUUGCUAAGCAGUGAUAAAUUUGGUCAAAGCCUAUUUGAUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGCAUUGAUUAAUCGUCGAUAACGAUAACCUG-----GCCAGUUAUGCAC ....(((...)))(((((((...((((((............(((((((((((((((((.....))))))))).))))))))(((((.....)))))....))-----)))).)))).))) ( -27.40) >DroSim_CAF1 20392 115 + 1 AAAUUGCUAAGCAGUGAUAAAUUUGGUCAAAGCCUAUUUGAUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGCAUUGAUUAAUCGUCGAUAACGAUAACCUG-----GCCAGUUAUGCAC ....(((...)))(((((((...((((((............(((((((((((((((((.....))))))))).))))))))(((((.....)))))....))-----)))).)))).))) ( -27.40) >DroEre_CAF1 20683 111 + 1 AAAUUGCUAAGCAGUGAUAAAUUUGGGCAAAGCCUAUUUGGUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCAUCGAUAACGAUAACCUG-----GCCAGUUA----U ..(((((...)))))........(((((...))))).(((((((((((((((((((((.....))))))))).)))))))....((((....))))......-----)))))...----. ( -25.10) >DroYak_CAF1 23616 120 + 1 AAAUUGCUAAGCAGUGAUAAAUUUGGCCAAAGCCUAUUUGAUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCAUCGAUAACGAUAAUAUGUUAUAGCCAGCUACGCAC ....(((..(((.(((((((((..(((....))).))))..(((((((((((((((((.....))))))))).)))))))))))))..((((((......)))))).....)))..))). ( -27.10) >consensus AAAUUGCUAAGCAGUGAUAAAUUUGGUCAAAGCCUAUUUGAUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCGUCGAUAACGAUAACCUG_____GCCAGUUAUGCAC ..(((((...))))).......(((((............(((((((((((((((((((.....))))))))).))))))).)))((((....))))...........)))))........ (-19.74 = -19.66 + -0.08)


| Location | 11,912,577 – 11,912,692 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.710850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
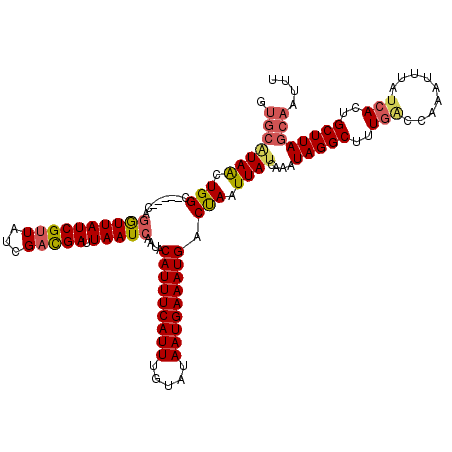
>X_DroMel_CAF1 11912577 115 - 22224390 AUGCAUAACUGGC-----CAGGUUAUCGUUAUCGCCGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACCAAUUAUCAAAUAGGCUUUGAUCAAAUUUAUAACUGCUUAGCAAUUU .(((((((.(((.-----..((((((((.......))).)))))....(((((((((.....))))))))).))).))))....(((((..(((......))).....)))))))).... ( -22.10) >DroSec_CAF1 22434 115 - 1 GUGCAUAACUGGC-----CAGGUUAUCGUUAUCGACGAUUAAUCAAUGCAUUUCAUUUGUAUAAUGAAAUGACUAAUUAUCAAAUAGGCUUUGACCAAAUUUAUCACUGCUUAGCAAUUU .(((((((.(((.-----..((((((((((...))))).)))))....(((((((((.....))))))))).))).))))....(((((..(((.........)))..)))))))).... ( -23.90) >DroSim_CAF1 20392 115 - 1 GUGCAUAACUGGC-----CAGGUUAUCGUUAUCGACGAUUAAUCAAUGCAUUUCAUUUGUAUAAUGAAAUGACUAAUUAUCAAAUAGGCUUUGACCAAAUUUAUCACUGCUUAGCAAUUU .(((((((.(((.-----..((((((((((...))))).)))))....(((((((((.....))))))))).))).))))....(((((..(((.........)))..)))))))).... ( -23.90) >DroEre_CAF1 20683 111 - 1 A----UAACUGGC-----CAGGUUAUCGUUAUCGAUGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACUAAUUACCAAAUAGGCUUUGCCCAAAUUUAUCACUGCUUAGCAAUUU .----.....(((-----(.((((((((....))))))))........(((((((((.....)))))))))...............))))((((...................))))... ( -21.21) >DroYak_CAF1 23616 120 - 1 GUGCGUAGCUGGCUAUAACAUAUUAUCGUUAUCGAUGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACUAAUUAUCAAAUAGGCUUUGGCCAAAUUUAUCACUGCUUAGCAAUUU .(((..(((((((((...(..(((((((....))))))).........(((((((((.....)))))))))...............)....))))))...........)))..))).... ( -24.90) >consensus GUGCAUAACUGGC_____CAGGUUAUCGUUAUCGACGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACUAAUUAUCAAAUAGGCUUUGACCAAAUUUAUCACUGCUUAGCAAUUU .(((((((.(((........((((((((((...))))).)))))....(((((((((.....))))))))).))).))))....(((((..(((.........)))..)))))))).... (-18.04 = -18.68 + 0.64)

| Location | 11,912,617 – 11,912,732 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -15.84 |
| Energy contribution | -15.48 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
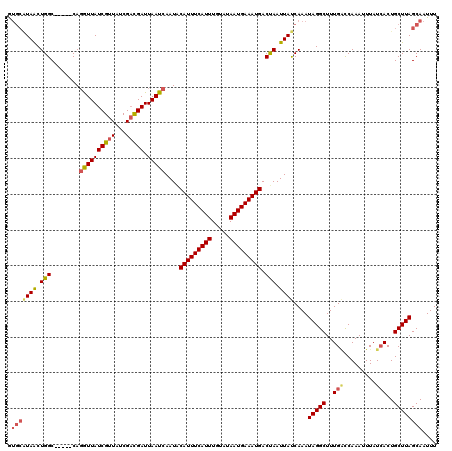
>X_DroMel_CAF1 11912617 115 + 22224390 AUAAUUGGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCGGCGAUAACGAUAACCUG-----GCCAGUUAUGCAUUCGACGAGAUAUAUUCAAAAGUCGGACACAAAUGCUGCAU .((((..(((((((((((.....))))))))).))..))))((((.......))))......-----((.(((..((..((((((..((.....))....))))))..))...))))).. ( -27.40) >DroSec_CAF1 22474 93 + 1 AUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGCAUUGAUUAAUCGUCGAUAACGAUAACCUG-----GCCAGUUAUGCACUCGA----------------------CACAAAUGCUGCAU .(((((((((((((((((.....))))))))).))))))))...(((((.....(((((...-----....)))))....))))----------------------)............. ( -20.90) >DroSim_CAF1 20432 93 + 1 AUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGCAUUGAUUAAUCGUCGAUAACGAUAACCUG-----GCCAGUUAUGCACUCGA----------------------CACAAAUGCUGCAU .(((((((((((((((((.....))))))))).))))))))...(((((.....(((((...-----....)))))....))))----------------------)............. ( -20.90) >DroEre_CAF1 20723 111 + 1 GUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCAUCGAUAACGAUAACCUG-----GCCAGUUA----UUCAAAGAGAUAUAUUAAACAGUCGGGCACAAAUGCUGCAU .(((((((((((((((((.....))))))))).))))))))...((((....))))..((((-----((...(((----.((.....)).....)))...)))))).............. ( -21.60) >DroYak_CAF1 23656 107 + 1 AUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCAUCGAUAACGAUAAUAUGUUAUAGCCAGCUACGCACUCAAUGAGAUAUAUU-------------CAAAUGCUGCAU .(((((((((((((((((.....))))))))).))))))))...((((....))))...........((.(((......................-------------.....))))).. ( -18.63) >consensus AUAAUUAGUCAUUUCAUUAUACAAAUGAAAUGUAUUGAUUAAUCGUCGAUAACGAUAACCUG_____GCCAGUUAUGCACUCGA_GAGAUAUAUU___________CACAAAUGCUGCAU .(((((((((((((((((.....))))))))).))))))))...((((....))))...........((.(((........................................))))).. (-15.84 = -15.48 + -0.36)



| Location | 11,912,617 – 11,912,732 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.55 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
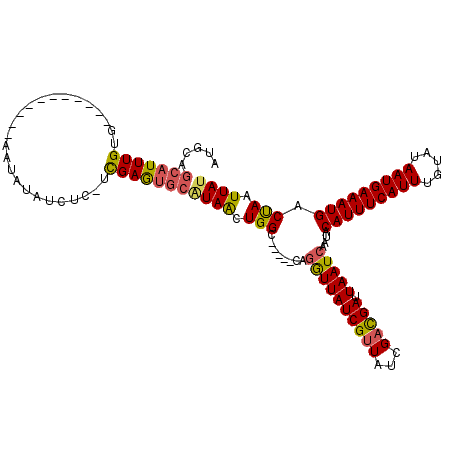
>X_DroMel_CAF1 11912617 115 - 22224390 AUGCAGCAUUUGUGUCCGACUUUUGAAUAUAUCUCGUCGAAUGCAUAACUGGC-----CAGGUUAUCGUUAUCGCCGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACCAAUUAU .....(((((((....(((..(........)..))).)))))))((((.(((.-----..((((((((.......))).)))))....(((((((((.....))))))))).))).)))) ( -23.70) >DroSec_CAF1 22474 93 - 1 AUGCAGCAUUUGUG----------------------UCGAGUGCAUAACUGGC-----CAGGUUAUCGUUAUCGACGAUUAAUCAAUGCAUUUCAUUUGUAUAAUGAAAUGACUAAUUAU ..(((..(((..((----------------------((((..((((((((...-----..)))))).))..))))))...)))...)))((((((((.....)))))))).......... ( -21.70) >DroSim_CAF1 20432 93 - 1 AUGCAGCAUUUGUG----------------------UCGAGUGCAUAACUGGC-----CAGGUUAUCGUUAUCGACGAUUAAUCAAUGCAUUUCAUUUGUAUAAUGAAAUGACUAAUUAU ..(((..(((..((----------------------((((..((((((((...-----..)))))).))..))))))...)))...)))((((((((.....)))))))).......... ( -21.70) >DroEre_CAF1 20723 111 - 1 AUGCAGCAUUUGUGCCCGACUGUUUAAUAUAUCUCUUUGAA----UAACUGGC-----CAGGUUAUCGUUAUCGAUGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACUAAUUAC ..........((.(((.(..(((((((.........)))))----)).).)))-----))((((((((....))))))))........(((((((((.....)))))))))......... ( -21.40) >DroYak_CAF1 23656 107 - 1 AUGCAGCAUUUG-------------AAUAUAUCUCAUUGAGUGCGUAGCUGGCUAUAACAUAUUAUCGUUAUCGAUGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACUAAUUAU .....(((((..-------------(..........)..)))))((((....)))).....(((((((....))))))).........(((((((((.....)))))))))......... ( -21.50) >consensus AUGCAGCAUUUGUG___________AAUAUAUCUC_UCGAGUGCAUAACUGGC_____CAGGUUAUCGUUAUCGACGAUUAAUCAAUACAUUUCAUUUGUAUAAUGAAAUGACUAAUUAU .....(((((((.........................)))))))((((.(((........((((((((((...))))).)))))....(((((((((.....))))))))).))).)))) (-16.67 = -16.55 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:03 2006