
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,909,949 – 11,910,109 |
| Length | 160 |
| Max. P | 0.993162 |

| Location | 11,909,949 – 11,910,046 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.31 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -12.68 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
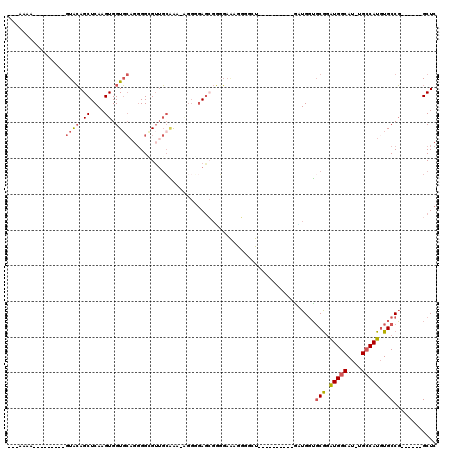
>X_DroMel_CAF1 11909949 97 + 22224390 ---AAAA---------GUACAGCUCAAGUGGUGCAGGGGCGUUGCAAAAAGGGGAGCGUGGAAAGGGGAU----GGCGGUGGUGGUGCGGAUGGCAU-UGCCAUGUGCCG------GCUC ---....---------.(((.((((...(..(((((.....)))))..)....))))))).....(((.(----((((((((..((((.....))))-..)))).)))))------.))) ( -33.70) >DroSec_CAF1 20387 94 + 1 AAAAAAA---------GUACAGCUCAAGUGGUGCAGGGGAGUUGCAAAAAGGGGAGCGGGGAAAGGGGCU----------GCUGGUGCGGAUGGCAU-UGCCAUGUGCCG------GCUC .......---------.....((((...(..(((((.....)))))..)....)))).............----------(((((..(..(((((..-.))))))..)))------)).. ( -28.20) >DroSim_CAF1 17899 94 + 1 AAAAAAA---------GUACAGCUCAAGUGGUGCAGGGGCGUUGCAAAAAGGGGAGCGGGGAAAGGGGCU----------GCUGGUGCGGAUGGCAU-UGCCAUGUGCCG------GCUC .......---------....(((((..((..(((....)))..))......(....)........)))))----------(((((..(..(((((..-.))))))..)))------)).. ( -29.10) >DroEre_CAF1 18316 76 + 1 ---AAAA---------GUACAGCUCAAGUGGUGCAGGGGCGUUGCAAA------AAGGGGG-------------------AAUGGUGCGGAUGGCAU-AGCCAUGUGCCG------GCUC ---....---------((((..(((...(..(((((.....)))))..------)...)))-------------------....))))((.((((((-(....)))))))------.)). ( -21.30) >DroYak_CAF1 21102 99 + 1 ---AAAA---------GUACAGCUCAAGUGGUGCAGGGGGGGGGGGGA--GGCGAGGGGCGAAGGUAGGUGGUAGCCGGUGUUGGUGCGGAUGGCAU-UGCCAUUUGCCG------GCUC ---....---------((((.((....)).))))..............--......((((...(((((((((((((((.(((....)))..))))..-))))))))))).------)))) ( -30.30) >DroAna_CAF1 40477 107 + 1 AGAAAAACUGAAACCCAUGAAGCUCAAGUGGUU----GGAGUUUCCA--GGGUGAGCCUUUAGUGGAGCC----UGC---CAUGUGCCAUGUGGCAUGUGGCAUGUGCCUGCUACGGCUC ............((((..(((((((........----.)))))))..--))))(((((..(((..(.(((----(((---((((((((....))))))))))).).)))..))).))))) ( -48.40) >consensus ___AAAA_________GUACAGCUCAAGUGGUGCAGGGGCGUUGCAAA_AGGGGAGCGGGGAAAGGGGCU__________GAUGGUGCGGAUGGCAU_UGCCAUGUGCCG______GCUC ................((((.((....)).))))....................................................(((.(((((....))))).)))............ (-12.68 = -13.10 + 0.42)


| Location | 11,910,013 – 11,910,109 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 86.18 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -28.53 |
| Energy contribution | -28.77 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
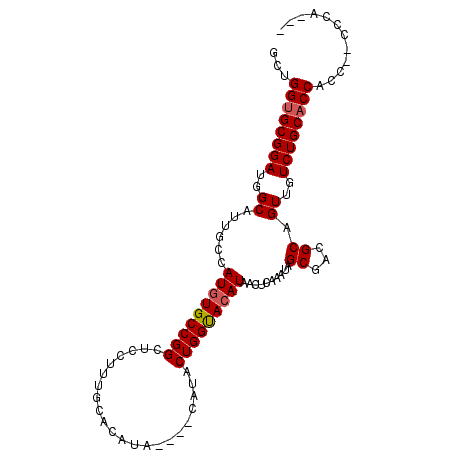
>X_DroMel_CAF1 11910013 96 + 22224390 GGUGGUGCGGAUGGCAUUGCCAUGUGCCGGCUCCUUUGCACAUA----CAUACUGGUACAUAACUCAAAUAGCGACGCAGUUGUCUGCACCACCCCCCCA--- (((((((((((..((......(((((((((..............----....)))))))))..........((...)).))..)))))))))))......--- ( -37.37) >DroSec_CAF1 20448 98 + 1 GCUGGUGCGGAUGGCAUUGCCAUGUGCCGGCUCCUUUGCACAUACAUACAUACUGGCACAUAACUCAAAUAGCGACGCAGUUGUCUGCACCACC--CCCA--- ..(((((((((..((......(((((((((......((......))......)))))))))..........((...)).))..)))))))))..--....--- ( -34.30) >DroSim_CAF1 17960 98 + 1 GCUGGUGCGGAUGGCAUUGCCAUGUGCCGGCUCCUUUGCACAUACAUACAUACUGGUACAUAACUCAAAUAGCGACGCAGUUGUCUGCACCACC--CCCA--- ..(((((((((..((......(((((((((......((......))......)))))))))..........((...)).))..)))))))))..--....--- ( -32.30) >DroEre_CAF1 18359 84 + 1 AAUGGUGCGGAUGGCAUAGCCAUGUGCCGGCUCCU-UGCAC--------AUACUGGUACAUAACUCAAAUAGCGACGCAGUUGUCUGCCCCCU---------- ...((.(((((..((......(((((((((.....-.....--------...)))))))))..........((...)).))..)))))))...---------- ( -25.32) >DroYak_CAF1 21168 93 + 1 GUUGGUGCGGAUGGCAUUGCCAUUUGCCGGCUCCUUUGCAC--------AUACUGGUACAUAACUCAAAUAGCGACGCAGUUGUCUGCACCCUC--CCCCUCC ...((((((((..((..(((((..((...((......)).)--------)...))))).............((...)).))..))))))))...--....... ( -26.20) >consensus GCUGGUGCGGAUGGCAUUGCCAUGUGCCGGCUCCUUUGCACAUA____CAUACUGGUACAUAACUCAAAUAGCGACGCAGUUGUCUGCACCACC__CCCA___ ...((((((((..((......(((((((((......................)))))))))..........((...)).))..))))))))............ (-28.53 = -28.77 + 0.24)

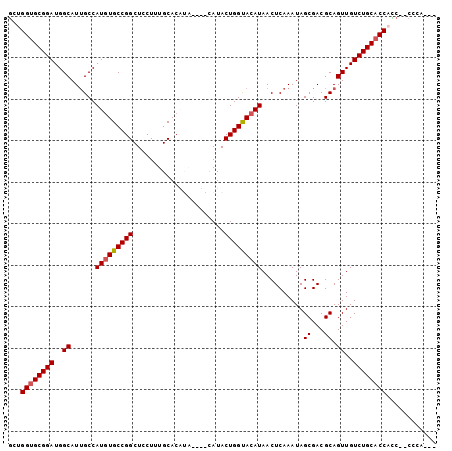
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:58 2006