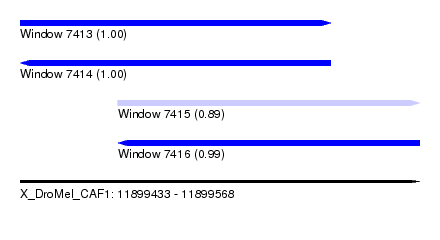
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,899,433 – 11,899,568 |
| Length | 135 |
| Max. P | 0.999281 |

| Location | 11,899,433 – 11,899,538 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.03 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -14.10 |
| Energy contribution | -17.29 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11899433 105 + 22224390 UAAACAGGAGCAAAAAA-GCGCUGAGG-GGAAAGUGCAUUGGGCUGGCUCCAGUGGAGGCCAAAAGGCAGAUAUAGCUGA--------AGCUGGAGCCGGAACUGGAGUCCCUGA ....((((..(......-(((((....-....)))))...(..(((((((((((....(((....)))((......))..--------.)))))))))))..)....)..)))). ( -42.00) >DroVir_CAF1 1536 86 + 1 UAAACACAAGCAAA-AA-GCGUUAAG--GGAAAGUGCAUUGGA-UGAC--AGGGUGCUGG-AAAAAGCAC----AGACCG------------GCAGCAGGACAUGGAGUG----- ....(((..((...-..-))......--.....(((..(((..-((..--.(((((((..-....)))))----...)).------------.)).)))..)))...)))----- ( -15.70) >DroSec_CAF1 10305 102 + 1 UAAACAGAAGCAAAAAACGCGUUGAGG-GGAAAGUGCAUUGGGCUGGCUCCGGUUGAGGCCAGAAGGCAGAUAUAGCCGA------------GGAGCCGGAACUGGAGUUCCUGA .........(((.....(.(.....).-).....))).(..((..((((((((((..((((....(((.......)))..------------.).)))..))))))))))))..) ( -33.00) >DroEre_CAF1 7760 85 + 1 UAAACAGGAGCAAA-AC-GCGCUGAGG-GGAAAGUGCAUUGGGCUGGCUCCGGUUGAGGCCAGAAGGCAGUUAUAG---------------------------UGGAGUCCCUGA ....(((((((..(-(.-(((((....-....))))).))..)))(((((((.((((.(((....)))..))).).---------------------------))))))))))). ( -31.80) >DroYak_CAF1 10009 114 + 1 UAAACAGGAGCAAAAAA-GCAAUGAGGGGGAAAGUGCAUUGGGCUGGCUCCAGUUGAGGCCAGAAGGCAGAUAUAGCCGAGGAGCAGGAGCCAAAGCCGGAACUGGAGCCCCUGA ....(((((((...((.-(((.............))).))..)))((((((((((..(((.....(((.......)))..((........))...)))..)))))))))))))). ( -40.42) >consensus UAAACAGGAGCAAAAAA_GCGCUGAGG_GGAAAGUGCAUUGGGCUGGCUCCGGUUGAGGCCAGAAGGCAGAUAUAGCCGA____________GGAGCCGGAACUGGAGUCCCUGA ....(((((((....((.(((((.........))))).))..)))((((((((((...(((....)))........(((..................))))))))))))))))). (-14.10 = -17.29 + 3.19)

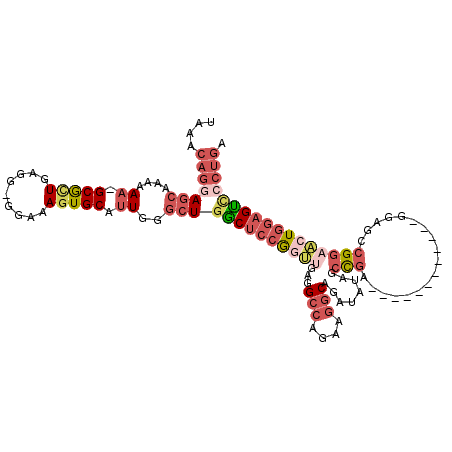
| Location | 11,899,433 – 11,899,538 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.03 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -6.74 |
| Energy contribution | -8.18 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.26 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11899433 105 - 22224390 UCAGGGACUCCAGUUCCGGCUCCAGCU--------UCAGCUAUAUCUGCCUUUUGGCCUCCACUGGAGCCAGCCCAAUGCACUUUCC-CCUCAGCGC-UUUUUUGCUCCUGUUUA ...(((((....)))))(((((((((.--------...)))......(((....))).......)))))).((.....)).......-...(((.((-......))..))).... ( -29.40) >DroVir_CAF1 1536 86 - 1 -----CACUCCAUGUCCUGCUGC------------CGGUCU----GUGCUUUUU-CCAGCACCCU--GUCA-UCCAAUGCACUUUCC--CUUAACGC-UU-UUUGCUUGUGUUUA -----(((....(((..((.((.------------(((...----(((((....-..))))).))--).))-..))..)))......--......((-..-...))..))).... ( -11.80) >DroSec_CAF1 10305 102 - 1 UCAGGAACUCCAGUUCCGGCUCC------------UCGGCUAUAUCUGCCUUCUGGCCUCAACCGGAGCCAGCCCAAUGCACUUUCC-CCUCAACGCGUUUUUUGCUUCUGUUUA ...(((((....)))))((((((------------..(((.......)))....((......))))))))(((..(((((.......-.......)))))....)))........ ( -28.64) >DroEre_CAF1 7760 85 - 1 UCAGGGACUCCA---------------------------CUAUAACUGCCUUCUGGCCUCAACCGGAGCCAGCCCAAUGCACUUUCC-CCUCAGCGC-GU-UUUGCUCCUGUUUA .(((((.((((.---------------------------........(((....))).......))))))(((..(((((.((....-....)).))-))-)..))).))).... ( -20.79) >DroYak_CAF1 10009 114 - 1 UCAGGGGCUCCAGUUCCGGCUUUGGCUCCUGCUCCUCGGCUAUAUCUGCCUUCUGGCCUCAACUGGAGCCAGCCCAAUGCACUUUCCCCCUCAUUGC-UUUUUUGCUCCUGUUUA .((((((((((((((..((((..(((....(((....))).......)))....))))..))))))))))(((.....(((.............)))-......))))))).... ( -38.82) >consensus UCAGGGACUCCAGUUCCGGCUCC____________UCGGCUAUAUCUGCCUUCUGGCCUCAACCGGAGCCAGCCCAAUGCACUUUCC_CCUCAACGC_UUUUUUGCUCCUGUUUA .(((((........................................(((...(((((..(....)..)))))......)))..............((.......))))))).... ( -6.74 = -8.18 + 1.44)


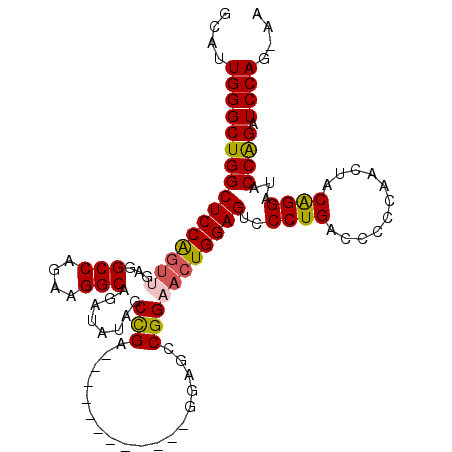
| Location | 11,899,466 – 11,899,568 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -36.59 |
| Consensus MFE | -24.44 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11899466 102 + 22224390 GCAUUGGGCUGGCUCCAGUGGAGGCCAAAAGGCAGAUAUAGCUGA--------AGCUGGAGCCGGAACUGGAGUCCCUGACCCCCAACUACGGGAUACCAGAUCCAGUAA ..(((((((((((((((((....(((....)))((......))..--------.)))))))))))..((((.(((((..............))))).)))).)))))).. ( -42.74) >DroSec_CAF1 10339 98 + 1 GCAUUGGGCUGGCUCCGGUUGAGGCCAGAAGGCAGAUAUAGCCGA------------GGAGCCGGAACUGGAGUUCCUGACCCCCAACUACGGGAUACCAGAUCCAGAAA ...((((((((((((((((....)))....(((.......)))..------------))))))))..((((...(((((...........)))))..)))).)))))... ( -37.90) >DroEre_CAF1 7792 79 + 1 GCAUUGGGCUGGCUCCGGUUGAGGCCAGAAGGCAGUUAUAG---------------------------UGGAGUCCCUGACCCCCAACUGCAGGAUACCGGAUCCA---- (((((((((((((..(....)..)))))..(((((..((..---------------------------....))..))).))))))).))).((((.....)))).---- ( -27.20) >DroYak_CAF1 10043 109 + 1 GCAUUGGGCUGGCUCCAGUUGAGGCCAGAAGGCAGAUAUAGCCGAGGAGCAGGAGCCAAAGCCGGAACUGGAGCCCCUGACCCCCAACUACAGGACACCAGAUCCAU-AU ......((((((((((.(((...(((....))).)))...((......)).))))))..))))(((.((((.(..((((...........)))).).)))).)))..-.. ( -38.50) >consensus GCAUUGGGCUGGCUCCAGUUGAGGCCAGAAGGCAGAUAUAGCCGA____________GGAGCCGGAACUGGAGUCCCUGACCCCCAACUACAGGAUACCAGAUCCAG_AA ....((((((((((((((((...(((....)))........(((..................)))))))))))..((((...........))))...)))).)))).... (-24.44 = -24.82 + 0.38)

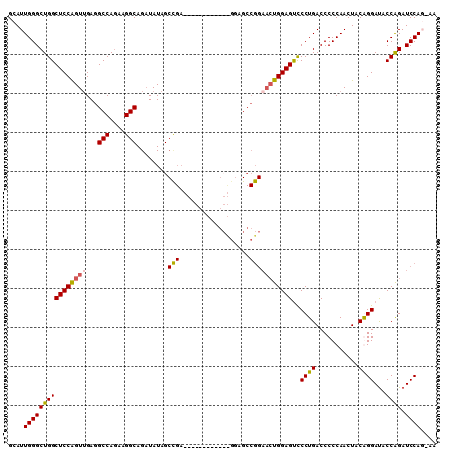
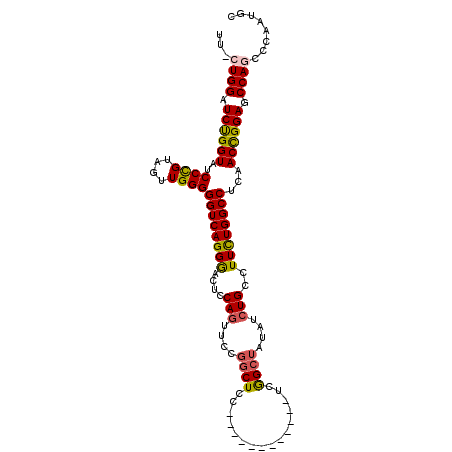
| Location | 11,899,466 – 11,899,568 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11899466 102 - 22224390 UUACUGGAUCUGGUAUCCCGUAGUUGGGGGUCAGGGACUCCAGUUCCGGCUCCAGCU--------UCAGCUAUAUCUGCCUUUUGGCCUCCACUGGAGCCAGCCCAAUGC ....(((..(((((.(((...(((.(((((((((((((....)))))(((...(((.--------...)))......)))...))))))))))))))))))).))).... ( -39.40) >DroSec_CAF1 10339 98 - 1 UUUCUGGAUCUGGUAUCCCGUAGUUGGGGGUCAGGAACUCCAGUUCCGGCUCC------------UCGGCUAUAUCUGCCUUCUGGCCUCAACCGGAGCCAGCCCAAUGC ...((((.((((((.....(((((((((((((.(((((....)))))))).))------------))))))))....(((....)))....)))))).))))........ ( -41.70) >DroEre_CAF1 7792 79 - 1 ----UGGAUCCGGUAUCCUGCAGUUGGGGGUCAGGGACUCCA---------------------------CUAUAACUGCCUUCUGGCCUCAACCGGAGCCAGCCCAAUGC ----(((.((((((..((.(((((((((((((...)))))).---------------------------...))))))).....)).....)))))).)))......... ( -27.20) >DroYak_CAF1 10043 109 - 1 AU-AUGGAUCUGGUGUCCUGUAGUUGGGGGUCAGGGGCUCCAGUUCCGGCUUUGGCUCCUGCUCCUCGGCUAUAUCUGCCUUCUGGCCUCAACUGGAGCCAGCCCAAUGC ..-.(((..(((((.((((((((((((((..((((((((..(((....)))..))))))))..)))))))))))...(((....))).......)))))))).))).... ( -48.30) >consensus UU_CUGGAUCUGGUAUCCCGUAGUUGGGGGUCAGGGACUCCAGUUCCGGCUCC____________UCGGCUAUAUCUGCCUUCUGGCCUCAACCGGAGCCAGCCCAAUGC ...((((.((((((..((((....))))((((((((....(((....((((................))))....)))..))))))))...)))))).))))........ (-25.15 = -25.34 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:53 2006