| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,894,292 – 11,894,399 |
| Length | 107 |
| Max. P | 0.782587 |

| Location | 11,894,292 – 11,894,399 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
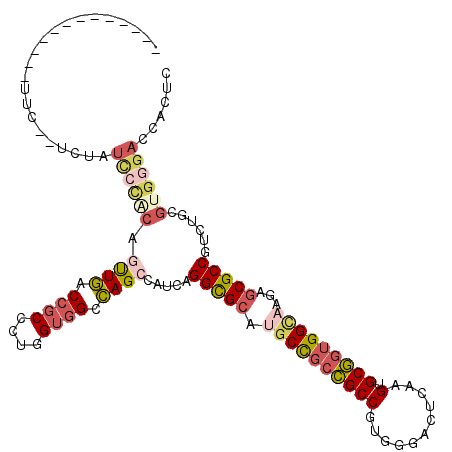
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -34.26 |
| Energy contribution | -35.73 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
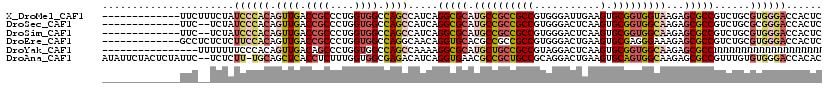
>X_DroMel_CAF1 11894292 107 + 22224390 -------------UUCUUUCUAUCCCACAGUUGACCGCCCUGGUGGCCAGCCAUCAGGCGCAUGCCGCCGCCGUGGGAUUGAAGUGCGGUGGUAAGAGCGCCGUCUGCGUGGGACCACUC -------------.........((((((.((.(((.....((((((....))))))(((((.((((((((((...........).)))))))))...)))))))).))))))))...... ( -48.20) >DroSec_CAF1 4964 105 + 1 -------------UUC--UCUAUCCCACAGUUGACCGCCCUGGUGGCCAGCCAUCAGGCGCAUGCCGCCGCCGUGGGACUCAAGUGCGGUGGCAAGAGCGCCGUCUGCGCGGGACCACUC -------------...--....((((..........((((((((((....)))))))).)).((((((((((.((.....)).).)))))))))...((((.....))))))))...... ( -48.60) >DroSim_CAF1 5615 105 + 1 -------------UUC--UCUAUCCCACAGUUGACCGCCCUGGUGGCCAGCCAUCAGGCGCAUGCCGCCGCCGUGGGACUCAAGUGCGGUGGCAAGAGCGCCGUCUGCGUGGGACCACUC -------------...--....((((((.((.(((.....((((((....))))))(((((.((((((((((.((.....)).).)))))))))...)))))))).))))))))...... ( -51.20) >DroEre_CAF1 5168 107 + 1 -------------GCCUCUCUCUUCCACAGUUGACCGCCCUGGUGGCCAGGCAACAGGUGCACGCCGCCGCCGUGGGACUGAAGUGCGAGGGAAAGAGCGCCGUCUGCGUGGGACCACUC -------------((((((.(((..(.((.((.(...((((((((((..(....).(((....)))))))))).)))..).)).)).)..))).)))).)).......(((....))).. ( -40.70) >DroYak_CAF1 5611 104 + 1 ----------------UUUUUUUCCCACAGUUGACAGCCCUGGUGGCCAGCCAAAAGGCGCAUGCUGCCGCCGUAGGACUCAAGUGCGGUGGCAAGAGCGCCNNNNNNNNNNNNNNNNNN ----------------.............((((...(((.....))))))).....(((((.(..((((((((((.........)))))))))).).))))).................. ( -30.10) >DroAna_CAF1 20833 117 + 1 AUAUUCUACUCUAUUC--UCUCUU-UGCAGCUCACCUCUUUGGUGGCGAGACAUCAGGUGAACGCCGCUGCCGCAGGACUGAAGUGCAGUGGCAAGAGCGCCGUUUGUGUGGGACCACAC ...((((((.......--......-....((.((((.....))))))...(((...((((...((((((((..((....))....)))))))).....))))...)))))))))...... ( -36.80) >consensus _____________UUC__UCUAUCCCACAGUUGACCGCCCUGGUGGCCAGCCAUCAGGCGCAUGCCGCCGCCGUGGGACUCAAGUGCGGUGGCAAGAGCGCCGUCUGCGUGGGACCACUC ......................((((((.((((.((((....)))).)))).....(((((.((((((((((...........).)))))))))...)))))......))))))...... (-34.26 = -35.73 + 1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:48 2006