
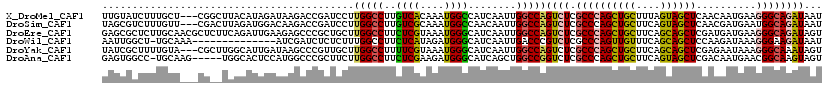
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,889,456 – 11,889,573 |
| Length | 117 |
| Max. P | 0.867027 |

| Location | 11,889,456 – 11,889,573 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.12 |
| Mean single sequence MFE | -38.60 |
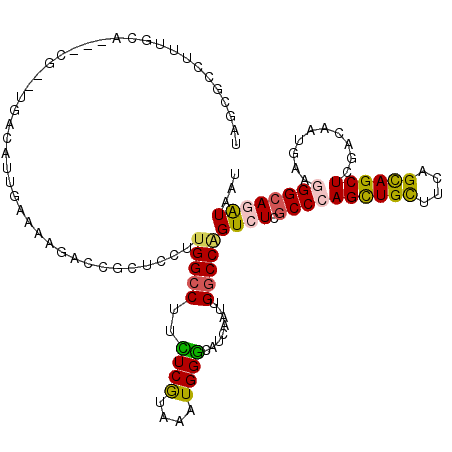
| Consensus MFE | -18.59 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11889456 117 + 22224390 UUGUAUCUUUGCU---CGGCUUACAUAGAUAAGACCGAUCCUUGGCCUUGUCACAAAUGGCCAUCAAUUGGCCAGUCUCGCCCAGCUGCUUUAGUAGCUCAACAAUGAAGGGCAGAUAAU ...(((((.....---.(((.......((((((.((((...)))).)))))).....((((((.....)))))))))..((((.(((((....)))))(((....))).))))))))).. ( -39.80) >DroSim_CAF1 777 117 + 1 UAGCGUCUUUGUU---CGACUUAGAUGGACAAGACCGAUCCUUGGCCUUGUCGCAAAUGGCCAACAAUUGGCCAGUCUCGCCCAGCUGCUUCAGUAGCUCAACGAUGAAUGGCAGAUAAU ....(((((..((---((((.....((((((((.((((...)))).)))))).))..((((((.....)))))))))(((...((((((....))))))...))).)))..).))))... ( -40.00) >DroEre_CAF1 734 120 + 1 GAGCGCUCUUGCAACGCUCUUCAGAUUGAAGAGCCCGCUGCUUGGCCUUCUCGUAAAUGGGCAUCAAUUGGCCAGUCUCGCCCAGCUGCUUCAGCAGCUCGAUGAUGAAGGGCAGAUAGU .(((((....))...(((((((.....)))))))..)))..(((.(((((.......(((.((.....)).)))((((((...((((((....))))))))).)))))))).)))..... ( -45.00) >DroWil_CAF1 184711 105 + 1 AAUUGGCU-UGCAAA--------------AUCGAUCUCUCUUUGGCCUUCUCAUAGAUGGGCAUCAAUUGACCCGUCUCGCCCAGUUGUUUCAGCAGCUCCAAGAUAAAGGGAAGAUAAU ........-......--------------....(((((((((((..(((.....(((((((((.....)).))))))).....((((((....))))))..))).))))))).))))... ( -25.80) >DroYak_CAF1 802 117 + 1 UAUCGCUUUUGUA---CGCUUGGCAUUGAUAAGCCCGUUGCUUGGCCUUUUCGUAAAUGGGCAUCAAUUGGCCAGUCUCGCCCAGCUGCUUCAGCAGCUCGAGAAUAAAGGGCAAAUAGU ....((((((...---...(((((((((((..(((((((...(((.....)))..)))))))))))))..)))))(((((...((((((....)))))))))))...))))))....... ( -40.60) >DroAna_CAF1 15354 114 + 1 GAGUGGCC-UGCAAG-----UGGCACUCCAUGGCCCGCUUCUUGGCCUUCUCGAAGAUGGGCAUCAGCUGGCCGGUCUCGCCCAGCUGCUUCAGUAGCUCGACAAUGAACGGCAAGUAGU ((.(((((-.((..(-----(((....)))).(((((((((..(.....)..)))).)))))....)).))))).))..(((..(((((....)))))((......))..)))....... ( -40.40) >consensus UAGCGCCUUUGCA___CG__UGACAUUGAAAAGACCGCUCCUUGGCCUUCUCGUAAAUGGGCAUCAAUUGGCCAGUCUCGCCCAGCUGCUUCAGCAGCUCGACAAUGAAGGGCAGAUAAU ..........................................(((((..((((....))))........)))))((((.((((((((((....))))))..........))))))))... (-18.59 = -18.23 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:45 2006