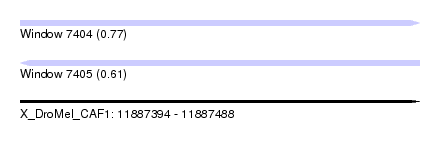
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,887,394 – 11,887,488 |
| Length | 94 |
| Max. P | 0.769179 |

| Location | 11,887,394 – 11,887,488 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.22 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11887394 94 + 22224390 CGAAGGGGAUUCCCCACCUCGGCCACGCCUU-CACUUCUUCCGCAAAGGACUUUCCCUUCGAUUCACUCGGAAAAAUCAUUAACUUAGCUACAUA (((((((((...........(((...)))..-.......(((.....)))..))))))))).................................. ( -19.00) >DroSec_CAF1 19299 92 + 1 CGAAGGGGAUUCCCCACCUCGGCCACGCCUU-CACUUUUUCCGCAGAGGACUUUCCUUUCCACUCACUCGGAAAAAUCAUUUGCGUAGCUACA-- ....((((...)))).....(((.((((...-...((((((((.((.(((........))).))....))))))))......)))).)))...-- ( -25.30) >DroEre_CAF1 20763 78 + 1 CGAAGGGGAUUCCCCACCUCGGCCACGCCUU-CACUUCUUCCGCAGAGGACUUUCC-----GCUCACUUGGAAAAAUCACU-G----------CG .(((((((....))).....(((...)))..-..))))...(((((.((..(((((-----(......))))))..)).))-)----------)) ( -23.30) >DroYak_CAF1 23392 90 + 1 CGAAGGGGAUUCCCCACCUCGGCCACGCCUUCCACUUCCUCCGCUGAGGACUUUCC-----ACACACCUGGAGAAAUCAUUUACGUUGCUACAUG ....((((...)))).....(((.(((.........(((((....))))).(((((-----(......)))))).........))).)))..... ( -22.40) >consensus CGAAGGGGAUUCCCCACCUCGGCCACGCCUU_CACUUCUUCCGCAGAGGACUUUCC_____ACUCACUCGGAAAAAUCAUUUACGUAGCUACAUG .(((((((....))).....(((...))).....)))).(((.....))).(((((.............)))))..................... (-14.60 = -14.22 + -0.38)



| Location | 11,887,394 – 11,887,488 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -19.80 |
| Energy contribution | -21.17 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11887394 94 - 22224390 UAUGUAGCUAAGUUAAUGAUUUUUCCGAGUGAAUCGAAGGGAAAGUCCUUUGCGGAAGAAGUG-AAGGCGUGGCCGAGGUGGGGAAUCCCCUUCG ......((((.(((.....((((((((.......(((((((....)))))))))))))))...-..))).))))(((((.(((....)))))))) ( -28.51) >DroSec_CAF1 19299 92 - 1 --UGUAGCUACGCAAAUGAUUUUUCCGAGUGAGUGGAAAGGAAAGUCCUCUGCGGAAAAAGUG-AAGGCGUGGCCGAGGUGGGGAAUCCCCUUCG --....(((((((......((((((((....((.(((........))).)).))))))))...-...)))))))(((((.(((....)))))))) ( -35.40) >DroEre_CAF1 20763 78 - 1 CG----------C-AGUGAUUUUUCCAAGUGAGC-----GGAAAGUCCUCUGCGGAAGAAGUG-AAGGCGUGGCCGAGGUGGGGAAUCCCCUUCG ((----------(-((.(..((((((........-----))))))..).))))).........-..(((...)))((((.(((....))))))). ( -26.60) >DroYak_CAF1 23392 90 - 1 CAUGUAGCAACGUAAAUGAUUUCUCCAGGUGUGU-----GGAAAGUCCUCAGCGGAGGAAGUGGAAGGCGUGGCCGAGGUGGGGAAUCCCCUUCG ......((.((((......(((((((.(.((...-----((.....)).)).)))))))).......)))).))(((((.(((....)))))))) ( -29.12) >consensus CAUGUAGCUACGCAAAUGAUUUUUCCAAGUGAGU_____GGAAAGUCCUCUGCGGAAGAAGUG_AAGGCGUGGCCGAGGUGGGGAAUCCCCUUCG ......(((((((......((((((((....((.....(((.....))))).)))))))).......)))))))(((((.(((....)))))))) (-19.80 = -21.17 + 1.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:43 2006