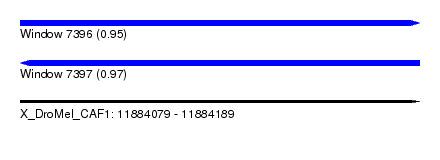
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,884,079 – 11,884,189 |
| Length | 110 |
| Max. P | 0.968478 |

| Location | 11,884,079 – 11,884,189 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11884079 110 + 22224390 CCCUUCUUUUUUU-CGCAUUUUGUUUCCCGGUUUUUUU----UUUCUUUUGAGAUGUCUUAUCAAAUCACCUUUUGCCUCUUCGUGUUGGGGCAAUGGCAAUAAUAAAAGGAAUG ...((((((((..-......(((((.((.(((......----((((....))))......)))..........(((((((........))))))).)).))))).)))))))).. ( -19.00) >DroSec_CAF1 15969 113 + 1 CCCUUCUAU-UUU-CGCAUUUUGGUUCCCCGUUUUAUUUUAUUUUUUUUUGAGAUGUCUUAUCAAAUCACCUUUUGCCUCUUCGUUUUGGGGCACUGGCAAUAAUAAAAGGAAUG .((((.(((-(..-.((..((((((....(((((((.............)))))))....))))))........((((((........))))))...))...)))).)))).... ( -22.92) >DroEre_CAF1 17486 105 + 1 CC-UUCUAU-UUUGUACGUUUUUUUUGU-----UUCU-CCACUUUUUGUUGAGAUGUCUUAUCAAAUCACCU-UUGCCUCUUCGU-UUGGGGCAAUGGCAAUAAUAAAAGGGACU ..-......-.......(((((((((((-----((((-(.((.....)).))))...............((.-(((((((.....-..))))))).))....)))))))))))). ( -25.00) >DroYak_CAF1 20442 103 + 1 CUGUUCUAU-UUU-UGCAUUUCUUUUCC-----UUUU----UUUUUUGUUGAGAUGUCUUAUCAAAUCACCUUUUGCCUCUUCGU-UUGGGGCAAUGGCAAUAAUAAAAGAAAUG .........-...-..((((((((((..-----....----...((((.((((....)))).))))...((..(((((((.....-..))))))).)).......)))))))))) ( -23.30) >consensus CCCUUCUAU_UUU_CGCAUUUUGUUUCC_____UUUU____UUUUUUGUUGAGAUGUCUUAUCAAAUCACCUUUUGCCUCUUCGU_UUGGGGCAAUGGCAAUAAUAAAAGGAAUG ................((((((((((..................((((.((((....)))).))))...((...((((((........))))))..)).......)))))))))) (-16.14 = -16.95 + 0.81)



| Location | 11,884,079 – 11,884,189 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11884079 110 - 22224390 CAUUCCUUUUAUUAUUGCCAUUGCCCCAACACGAAGAGGCAAAAGGUGAUUUGAUAAGACAUCUCAAAAGAAA----AAAAAAACCGGGAAACAAAAUGCG-AAAAAAAGAAGGG ....((((((....((((..(((((.(........).))))).(((((.(((....)))))))).........----..........(....).....)))-).....)))))). ( -17.70) >DroSec_CAF1 15969 113 - 1 CAUUCCUUUUAUUAUUGCCAGUGCCCCAAAACGAAGAGGCAAAAGGUGAUUUGAUAAGACAUCUCAAAAAAAAAUAAAAUAAAACGGGGAACCAAAAUGCG-AAA-AUAGAAGGG ....(((((((((.((((...((((.(........).))))..(((((.(((....)))))))).....................((....)).....)))-).)-)))))))). ( -24.70) >DroEre_CAF1 17486 105 - 1 AGUCCCUUUUAUUAUUGCCAUUGCCCCAA-ACGAAGAGGCAA-AGGUGAUUUGAUAAGACAUCUCAACAAAAAGUGG-AGAA-----ACAAAAAAAACGUACAAA-AUAGAA-GG ....(((((((((((..((.(((((.(..-.....).)))))-.))..))...........((((.((.....)).)-))).-----.................)-))))))-)) ( -24.20) >DroYak_CAF1 20442 103 - 1 CAUUUCUUUUAUUAUUGCCAUUGCCCCAA-ACGAAGAGGCAAAAGGUGAUUUGAUAAGACAUCUCAACAAAAAA----AAAA-----GGAAAAGAAAUGCA-AAA-AUAGAACAG ((((((((((....(..((.(((((.(..-.....).)))))..))..)((((...((....))...))))...----....-----..))))))))))..-...-......... ( -18.40) >consensus CAUUCCUUUUAUUAUUGCCAUUGCCCCAA_ACGAAGAGGCAAAAGGUGAUUUGAUAAGACAUCUCAAAAAAAAA____AAAA_____GGAAAAAAAAUGCA_AAA_AUAGAAGGG ...(((((((((...(((..(((((.(........).))))).(((((.(((....))))))))..................................))).....))))))))) (-14.32 = -14.32 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:36 2006