
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,882,821 – 11,882,937 |
| Length | 116 |
| Max. P | 0.910293 |

| Location | 11,882,821 – 11,882,937 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -19.66 |
| Energy contribution | -21.10 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
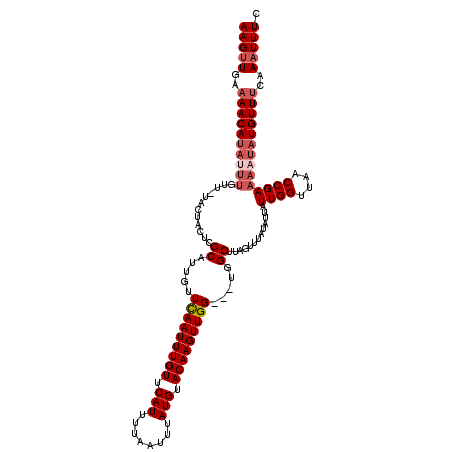
>X_DroMel_CAF1 11882821 116 + 22224390 AAGUUGAAAACAUAUUUGUU-CACUAGUCGCAUUGUUCAAUUUGUUCAUUUUAAUUUAUGUACAAGUUGG---UGGCUUAGUUUAUAUUAUUGGUUAACCGAAAAUAUGUUUCAAAUUUC ...((((((.(((((((...-.(((((((((......((((((((.(((........))).)))))))))---)))).))))........((((....)))))))))))))))))..... ( -27.10) >DroSec_CAF1 14713 116 + 1 AAGUUGAAAACAUUUUUGUU-UACUACUCGCAUUGUUCAAUUUGUUCAUUUUAAUUUAUGUACAAGUUGG---UGGCUUAGUUUAUAUUAUUGGUUAACCGAAAAUAUGUUUCAAAUUUC ...((((((...........-.((((...((.....(((((((((.(((........))).)))))))))---..)).)))).((((((.((((....)))).))))))))))))..... ( -21.70) >DroSim_CAF1 18671 116 + 1 AAGUUGAAAACAUAUUUGUU-UACUACUCGCAUUGUUCAAUUUGUUCAUUUUAAUUUAUGUACAAGUUGG---UGGCUUAGUUUAUAUUAUUGGUUAACCGAAAAUAUGUUUCAAAUUUC ...((((((.(((((((((.-.((((...((.....(((((((((.(((........))).)))))))))---..)).))))..))....((((....)))))))))))))))))..... ( -25.00) >DroEre_CAF1 16062 105 + 1 AAGUUGAAAACACAC------------UCGCAUUGUUCAAUUUGUUCAUUUUAAUUUAUGUACAAGUUGG---UGGCUUAGUUUAUAAUAUUGGUUAACCGAAAAUAUGUUCAAAAUUUC ...(((((.......------------..((.....(((((((((.(((........))).)))))))))---..)).............((((....)))).......)))))...... ( -16.90) >DroYak_CAF1 19176 120 + 1 AAGUUGAAAACAUACUUGUUUUUCUGCUCGCAUUGUUUAAUUUGUUCAUUUUGAUUUAUGUACAAGUUGGUGGUGGCUUAUUUUAUAAUAUUGGUUAACCGAAAAUAUGUUUAAACUUUU (((((..(((((((...........((.(((.....(((((((((.(((........))).)))))))))..))))).............((((....))))...))))))).))))).. ( -22.60) >consensus AAGUUGAAAACAUAUUUGUU_UACUACUCGCAUUGUUCAAUUUGUUCAUUUUAAUUUAUGUACAAGUUGG___UGGCUUAGUUUAUAUUAUUGGUUAACCGAAAAUAUGUUUCAAAUUUC (((((..((((((((((............((.....(((((((((.(((........))).))))))))).....)).............((((....))))))))))))))..))))). (-19.66 = -21.10 + 1.44)

| Location | 11,882,821 – 11,882,937 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -12.84 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11882821 116 - 22224390 GAAAUUUGAAACAUAUUUUCGGUUAACCAAUAAUAUAAACUAAGCCA---CCAACUUGUACAUAAAUUAAAAUGAACAAAUUGAACAAUGCGACUAGUG-AACAAAUAUGUUUUCAACUU .....(((((((((((((((((....))..........((((.((..---.(((.((((.(((........))).)))).)))......))...)))))-)..))))))).))))))... ( -17.30) >DroSec_CAF1 14713 116 - 1 GAAAUUUGAAACAUAUUUUCGGUUAACCAAUAAUAUAAACUAAGCCA---CCAACUUGUACAUAAAUUAAAAUGAACAAAUUGAACAAUGCGAGUAGUA-AACAAAAAUGUUUUCAACUU .....((((((.(((((((.((....))..........((((.((..---.(((.((((.(((........))).)))).)))......))...)))).-....)))))))))))))... ( -15.50) >DroSim_CAF1 18671 116 - 1 GAAAUUUGAAACAUAUUUUCGGUUAACCAAUAAUAUAAACUAAGCCA---CCAACUUGUACAUAAAUUAAAAUGAACAAAUUGAACAAUGCGAGUAGUA-AACAAAUAUGUUUUCAACUU .....(((((((((((((..((....))..........((((.((..---.(((.((((.(((........))).)))).)))......))...)))).-...))))))).))))))... ( -18.00) >DroEre_CAF1 16062 105 - 1 GAAAUUUUGAACAUAUUUUCGGUUAACCAAUAUUAUAAACUAAGCCA---CCAACUUGUACAUAAAUUAAAAUGAACAAAUUGAACAAUGCGA------------GUGUGUUUUCAACUU ........((((((((((.(((((..................)))).---.(((.((((.(((........))).)))).)))......).))------------))))))))....... ( -14.67) >DroYak_CAF1 19176 120 - 1 AAAAGUUUAAACAUAUUUUCGGUUAACCAAUAUUAUAAAAUAAGCCACCACCAACUUGUACAUAAAUCAAAAUGAACAAAUUAAACAAUGCGAGCAGAAAAACAAGUAUGUUUUCAACUU ..(((((.((((((((((..((....))...............((..........((((.(((........))).))))..........))............))))))))))..))))) ( -14.55) >consensus GAAAUUUGAAACAUAUUUUCGGUUAACCAAUAAUAUAAACUAAGCCA___CCAACUUGUACAUAAAUUAAAAUGAACAAAUUGAACAAUGCGAGUAGUA_AACAAAUAUGUUUUCAACUU .......(((((((((((..((....))...............((......(((.((((.(((........))).)))).)))......))............)))))))))))...... (-12.84 = -13.48 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:34 2006