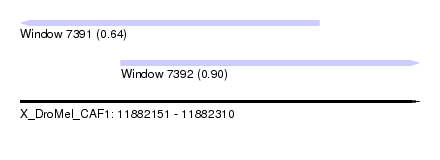
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,882,151 – 11,882,310 |
| Length | 159 |
| Max. P | 0.896281 |

| Location | 11,882,151 – 11,882,270 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11882151 119 - 22224390 AUUUCCGCCAUGCAAAAUCAACAGCAGCC-GACAACAAAUCCAGCCAACUGACCGGCUGUCAGGCAGCCAACCCACCUGGCCAUGUGCAAAUGCGUUUCACUGAAUCAUGUAUGGGUGCA .......................(((.((-((((.......(((((........)))))(((((((((((.......))))..)))((....))......))))....))).))).))). ( -29.50) >DroSec_CAF1 14032 120 - 1 AUUUCCGCCAUGCAAAAUCAACAGCAGCCGGACAACAAAUCCAGCCAACUGACCGGCUGUCAGGCAGCCAACCCACCUGGCCAUGUGCAAAUGCGUUUCACUGAAUCAUGCAUGGGUGCA .......(((((((..........(((((((.((...............)).)))))))(((((((((((.......))))..)))((....))......))))....)))))))..... ( -34.76) >DroSim_CAF1 16472 119 - 1 AUUUCCGCCAUGCAAAAUCAACAGCAGCCGGACAACAAAUCCAGCCAACUGACCGGCUGUCA-GCAGCCAACCCACCUGGCCAUGUGCAAAUGCGUUUCACUGAAUCAUGCAUGGGUGCA .......(((((((...((((((...(((((..........(((....)))...(((((...-.))))).......)))))..)))((....)).......)))....)))))))..... ( -33.60) >DroEre_CAF1 15396 119 - 1 AUUUCCGCCAUGCAAAAUCAACAGCAGCC-GACAACAAAUCCAACCAACUGACCGGCUGUCAGGCAGCCAACCCACCUGGCCAUGUGCAAAUGCGUUUCACUGAAUCAUGUAUGGGCACA ......((((((((..........(((((-(......................))))))(((((((((((.......))))..)))((....))......))))....))))).)))... ( -28.85) >DroYak_CAF1 18551 119 - 1 AUUUCCGCCAUGCAAAAUCAACAGCAGCC-GACAACAAAUCCAACCAGCUGACCGGCUGUCAGGCAGCCAACCCACCUGGCCAUGUGCAAAUGCGUUUUACUGAAUCAUGUAUGGGCACA ......((((...........((((....-.................))))...(((((.....)))))........))))..(((((..((((((...........))))))..))))) ( -28.90) >consensus AUUUCCGCCAUGCAAAAUCAACAGCAGCC_GACAACAAAUCCAGCCAACUGACCGGCUGUCAGGCAGCCAACCCACCUGGCCAUGUGCAAAUGCGUUUCACUGAAUCAUGUAUGGGUGCA .......(((((((...((((((...(((............(((....)))...(((((.....))))).........)))..)))((....)).......)))....)))))))..... (-26.74 = -26.90 + 0.16)

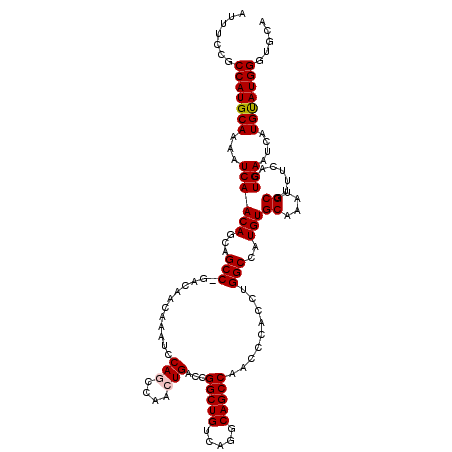
| Location | 11,882,191 – 11,882,310 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -33.02 |
| Energy contribution | -32.98 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11882191 119 + 22224390 CCAGGUGGGUUGGCUGCCUGACAGCCGGUCAGUUGGCUGGAUUUGUUGUC-GGCUGCUGUUGAUUUUGCAUGGCGGAAAUAUCAGCGACAAAGUUACCAAAAUAUUGAAAACAAAUAAAA ...(((((.(((((.(((.((((((..(((((....)).)))..))))))-))).)))(((((((((((...)))))...))))))....)).)))))...................... ( -34.30) >DroSec_CAF1 14072 120 + 1 CCAGGUGGGUUGGCUGCCUGACAGCCGGUCAGUUGGCUGGAUUUGUUGUCCGGCUGCUGUUGAUUUUGCAUGGCGGAAAUAUCAGCGACAAAGUUACCAAAAUAUUGAAAACAAAUAAAA ...(((((.(((((((.....))))).(((....((((((((.....))))))))((((.((.((((((...)))))).)).))))))).)).)))))...................... ( -39.00) >DroSim_CAF1 16512 119 + 1 CCAGGUGGGUUGGCUGC-UGACAGCCGGUCAGUUGGCUGGAUUUGUUGUCCGGCUGCUGUUGAUUUUGCAUGGCGGAAAUAUCAGCGACAAAGUUACCAAAAUAUUGAAAACAAAUAAAA ...(((((.(((((((.-...))))).(((....((((((((.....))))))))((((.((.((((((...)))))).)).))))))).)).)))))...................... ( -39.00) >DroEre_CAF1 15436 119 + 1 CCAGGUGGGUUGGCUGCCUGACAGCCGGUCAGUUGGUUGGAUUUGUUGUC-GGCUGCUGUUGAUUUUGCAUGGCGGAAAUAUCAGCGACAAAGUUACCAAAAUAUUGAAAACAAAUAAAA ((((.(.(..((((((.....))))))..)).))))((((.....(((((-(.(((.((((...(((((...))))))))).))))))))).....)))).................... ( -32.60) >DroYak_CAF1 18591 119 + 1 CCAGGUGGGUUGGCUGCCUGACAGCCGGUCAGCUGGUUGGAUUUGUUGUC-GGCUGCUGUUGAUUUUGCAUGGCGGAAAUAUCAGCGACAAAGUUACCAAAAUAUUGAAAACAAAUAAAA ((((.(.(..((((((.....))))))..)).))))((((.....(((((-(.(((.((((...(((((...))))))))).))))))))).....)))).................... ( -34.70) >consensus CCAGGUGGGUUGGCUGCCUGACAGCCGGUCAGUUGGCUGGAUUUGUUGUC_GGCUGCUGUUGAUUUUGCAUGGCGGAAAUAUCAGCGACAAAGUUACCAAAAUAUUGAAAACAAAUAAAA ...(((((.(((((.(((.((((((..(((((....)).)))..)))))).))).)))(((((((((((...)))))...))))))....)).)))))...................... (-33.02 = -32.98 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:32 2006